How to get a mesh from an MRI image?#
[1]:
import pyvista as pv
import polpo.preprocessing.dict as ppdict
from polpo.preprocessing.load.pregnancy.pilot import (
HippocampalSubfieldsSegmentationsLoader,
MriLoader,
)
from polpo.preprocessing.mesh.conversion import PvFromData
from polpo.preprocessing.mri import (
MeshExtractorFromSegmentedImage,
SkimageMarchingCubes,
)
[KeOps] Warning : cuda was detected, but driver API could not be initialized. Switching to cpu only.
[2]:
STATIC_VIZ = True
if STATIC_VIZ:
pv.set_jupyter_backend("static")
Loading data#
Following How to visualize MRI data?, we start by getting the image data.
[3]:
SESSION_ID = 1
[4]:
loader = MriLoader(subset=[SESSION_ID], as_image=True)
pipe = loader + ppdict.ExtractUniqueKey()
[5]:
img_fdata = pipe()
INFO: Data has already been downloaded... using cached file ('/home/luisfpereira/.herbrain/data/pregnancy/raw/mri').
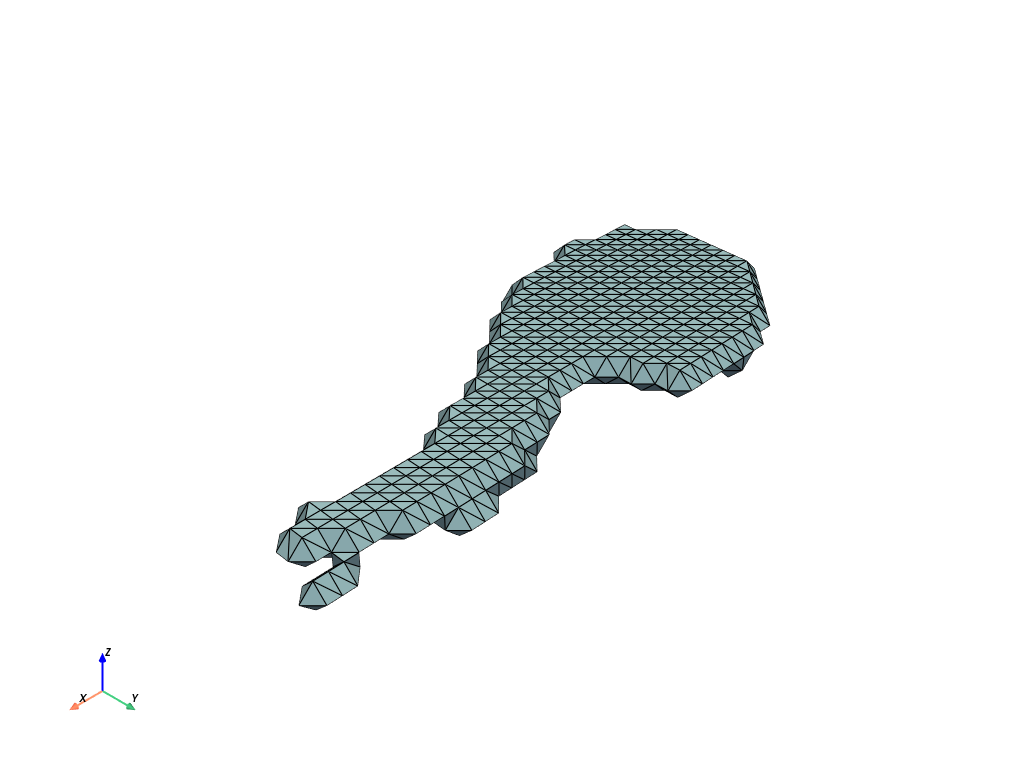
Marching cubes#
We now use marching cubes and transform the output to pyvista.PolyData for visualization.
[6]:
mesh_from_image = SkimageMarchingCubes()
[7]:
mesh = (mesh_from_image + PvFromData())(img_fdata)
Visualization#
[8]:
pl = pv.Plotter(border=False)
pl.show_axes()
pl.add_mesh(mesh, show_edges=True)
pl.show()
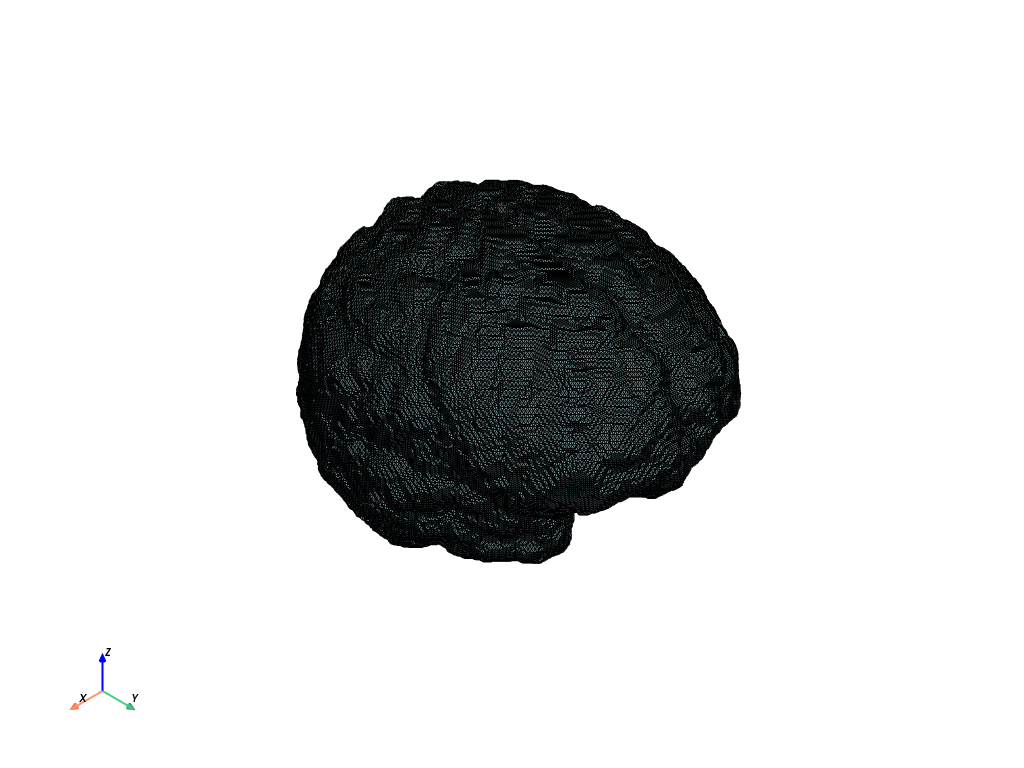
Segmented data#
Load the segmented data.
[9]:
pipe = (
HippocampalSubfieldsSegmentationsLoader(subset=[SESSION_ID], as_image=True)
+ ppdict.ExtractUniqueKey()
)
[10]:
img_fdata = pipe()
INFO: Data has already been downloaded... using cached file ('/home/luisfpereira/.herbrain/data/pregnancy/derivatives/segmentations').
The process changes only slightly when the data is segmented. (NB: encoding informs about the tool used for segmentation, which matters to map ids to tool ids and colors.)
[11]:
img2mesh = (
MeshExtractorFromSegmentedImage(
return_colors=True,
encoding="ashs",
)
+ PvFromData()
)
mesh = img2mesh(img_fdata)
[12]:
pl = pv.Plotter(border=False)
pl.show_axes()
pl.add_mesh(mesh, show_edges=True)
pl.show()
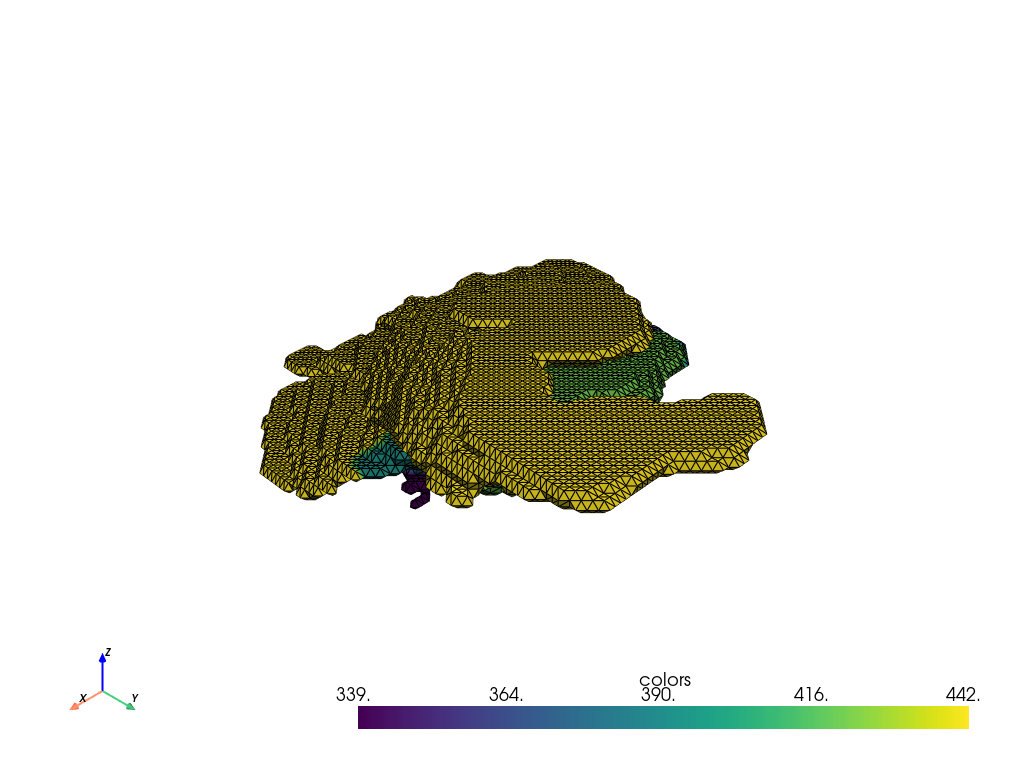
For example, we can select a particular subfield of the hippocampus.
[13]:
img2mesh = (
MeshExtractorFromSegmentedImage(
struct_id="PostHipp", return_colors=False, encoding="ashs"
)
+ PvFromData()
)
mesh = img2mesh(img_fdata)
[14]:
pl = pv.Plotter(border=False)
pl.show_axes()
pl.add_mesh(mesh, show_edges=True)
pl.show()