LDDMM: importance of the data attachment term#
[1]:
import shutil
from pathlib import Path
import numpy as np
import pyvista as pv
from matplotlib import pyplot as plt
import polpo.lddmm as plddmm
from polpo.mesh.qoi import centroid2farthest_vertex, vertexwise_euclidean
from polpo.mesh.surface import PvSurface
from polpo.mesh.varifold.tuning import SigmaFromLengths
from polpo.plot.pyvista import RegisteredMeshesColoredPlotter
from polpo.preprocessing.load.pregnancy.deformetrica import get_two_random_meshes
[KeOps] Warning : CUDA was detected, but driver API could not be initialized. Switching to CPU only.
[2]:
RECOMPUTE = False
STATIC_VIZ = True
VIZ = 1
if STATIC_VIZ:
pv.set_jupyter_backend("static")
[3]:
STRUCT_NAME = "L_Hipp"
OUTPUTS_DIR = Path.home() / ".polpo/results" / f"lddmm_data_attachment_{STRUCT_NAME}"
REGISTRATION_DIR = OUTPUTS_DIR / "registration"
if OUTPUTS_DIR.exists() and RECOMPUTE:
shutil.rmtree(OUTPUTS_DIR)
[4]:
meshes = get_two_random_meshes(
OUTPUTS_DIR,
mesh_names=("source", "target"),
target_reduction=0,
)
[5]:
if VIZ > 1:
pl = pv.Plotter(border=False)
for mesh in meshes:
pl.add_mesh(mesh.as_pv(), show_edges=True, opacity=0.6)
pl.show()
Following LDDMM: how to register a mesh to a template?.
[6]:
registration_kwargs = dict(
kernel_width=4.0,
regularisation=1.0,
max_iter=2000,
freeze_control_points=False,
tol=1e-16,
)
registration_dirs = []
mesh_filenames = list(meshes.keys())
[7]:
sigma_search = SigmaFromLengths(
ratio_charlen_mesh=2.0,
ratio_charlen=0.25,
)
sigma_search.fit(meshes.values())
varifold_metric = sigma_search.optimal_metric_
[8]:
metric = "landmark"
registration_dir = REGISTRATION_DIR / metric
registration_dirs.append(registration_dir)
if not registration_dir.exists():
plddmm.registration.estimate_registration(
mesh_filenames[0],
mesh_filenames[1],
metric=metric,
output_dir=registration_dir,
**registration_kwargs,
)
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_data_attachment_L_Hipp/registration/landmark/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -4.152E+02 [ attachment = -2.133E+02 ; regularity = -2.019E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -3.743E+02 [ attachment = -1.666E+02 ; regularity = -2.077E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -3.556E+02 [ attachment = -1.487E+02 ; regularity = -2.069E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -3.453E+02 [ attachment = -1.403E+02 ; regularity = -2.051E+02 ]
------------------------------------- Iteration: 100 -------------------------------------
>> Log-likelihood = -3.394E+02 [ attachment = -1.347E+02 ; regularity = -2.047E+02 ]
------------------------------------- Iteration: 120 -------------------------------------
>> Log-likelihood = -3.359E+02 [ attachment = -1.310E+02 ; regularity = -2.050E+02 ]
------------------------------------- Iteration: 140 -------------------------------------
>> Log-likelihood = -3.335E+02 [ attachment = -1.280E+02 ; regularity = -2.055E+02 ]
------------------------------------- Iteration: 160 -------------------------------------
>> Log-likelihood = -3.315E+02 [ attachment = -1.259E+02 ; regularity = -2.055E+02 ]
------------------------------------- Iteration: 180 -------------------------------------
>> Log-likelihood = -3.301E+02 [ attachment = -1.240E+02 ; regularity = -2.061E+02 ]
------------------------------------- Iteration: 200 -------------------------------------
>> Log-likelihood = -3.289E+02 [ attachment = -1.230E+02 ; regularity = -2.059E+02 ]
>> Gradient at Termination: 4.668068721272594
>> ABNORMAL:
>> Estimation took: 53 seconds
NB: use_svf=True affects the deformation model, not the data attachment term.
[9]:
metric = "landmark"
registration_dir = REGISTRATION_DIR / f"{metric}_svf"
registration_dirs.append(registration_dir)
if not registration_dir.exists():
plddmm.registration.estimate_registration(
mesh_filenames[0],
mesh_filenames[1],
use_svf=True,
metric=metric,
output_dir=registration_dir,
**registration_kwargs,
)
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_data_attachment_L_Hipp/registration/landmark_svf/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -4.341E+02 [ attachment = -2.262E+02 ; regularity = -2.078E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -3.891E+02 [ attachment = -1.757E+02 ; regularity = -2.134E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -3.724E+02 [ attachment = -1.611E+02 ; regularity = -2.113E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -3.633E+02 [ attachment = -1.525E+02 ; regularity = -2.108E+02 ]
------------------------------------- Iteration: 100 -------------------------------------
>> Log-likelihood = -3.582E+02 [ attachment = -1.479E+02 ; regularity = -2.103E+02 ]
------------------------------------- Iteration: 120 -------------------------------------
>> Log-likelihood = -3.547E+02 [ attachment = -1.432E+02 ; regularity = -2.114E+02 ]
------------------------------------- Iteration: 140 -------------------------------------
>> Log-likelihood = -3.522E+02 [ attachment = -1.405E+02 ; regularity = -2.118E+02 ]
------------------------------------- Iteration: 160 -------------------------------------
>> Log-likelihood = -3.504E+02 [ attachment = -1.384E+02 ; regularity = -2.120E+02 ]
------------------------------------- Iteration: 180 -------------------------------------
>> Log-likelihood = -3.490E+02 [ attachment = -1.367E+02 ; regularity = -2.123E+02 ]
------------------------------------- Iteration: 200 -------------------------------------
>> Log-likelihood = -3.479E+02 [ attachment = -1.354E+02 ; regularity = -2.125E+02 ]
>> Gradient at Termination: 39.79652326817482
>> CONVERGENCE: RELATIVE REDUCTION OF F <= FACTR*EPSMCH
>> Estimation took: 35 seconds
[10]:
metric = "varifold"
registration_dir = REGISTRATION_DIR / metric
registration_dirs.append(registration_dir)
if not registration_dir.exists():
plddmm.registration.estimate_registration(
mesh_filenames[0],
mesh_filenames[1],
metric=metric,
output_dir=registration_dir,
attachment_kernel_width=sigma_search.sigma_,
**registration_kwargs,
)
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_data_attachment_L_Hipp/registration/varifold/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -2.891E+02 [ attachment = -1.325E+02 ; regularity = -1.566E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -2.212E+02 [ attachment = -5.847E+01 ; regularity = -1.627E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -2.024E+02 [ attachment = -4.375E+01 ; regularity = -1.586E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -1.927E+02 [ attachment = -3.778E+01 ; regularity = -1.549E+02 ]
------------------------------------- Iteration: 100 -------------------------------------
>> Log-likelihood = -1.878E+02 [ attachment = -3.306E+01 ; regularity = -1.547E+02 ]
------------------------------------- Iteration: 120 -------------------------------------
>> Log-likelihood = -1.838E+02 [ attachment = -3.322E+01 ; regularity = -1.506E+02 ]
------------------------------------- Iteration: 140 -------------------------------------
>> Log-likelihood = -1.810E+02 [ attachment = -3.131E+01 ; regularity = -1.497E+02 ]
------------------------------------- Iteration: 160 -------------------------------------
>> Log-likelihood = -1.787E+02 [ attachment = -2.938E+01 ; regularity = -1.494E+02 ]
>> Gradient at Termination: 106.3541200889455
>> ABNORMAL:
>> Estimation took: 02 minutes and 32 seconds
[11]:
metric = "varifold"
registration_dir = REGISTRATION_DIR / f"{metric}_svf"
registration_dirs.append(registration_dir)
if not registration_dir.exists():
plddmm.registration.estimate_registration(
mesh_filenames[0],
mesh_filenames[1],
metric=metric,
use_svf=True,
output_dir=registration_dir,
attachment_kernel_width=sigma_search.sigma_,
**registration_kwargs,
)
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_data_attachment_L_Hipp/registration/varifold_svf/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -3.112E+02 [ attachment = -1.494E+02 ; regularity = -1.618E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -2.367E+02 [ attachment = -6.753E+01 ; regularity = -1.691E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -2.144E+02 [ attachment = -4.800E+01 ; regularity = -1.664E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -2.043E+02 [ attachment = -4.031E+01 ; regularity = -1.639E+02 ]
------------------------------------- Iteration: 100 -------------------------------------
>> Log-likelihood = -1.990E+02 [ attachment = -3.762E+01 ; regularity = -1.614E+02 ]
------------------------------------- Iteration: 120 -------------------------------------
>> Log-likelihood = -1.948E+02 [ attachment = -3.419E+01 ; regularity = -1.606E+02 ]
------------------------------------- Iteration: 140 -------------------------------------
>> Log-likelihood = -1.918E+02 [ attachment = -3.225E+01 ; regularity = -1.595E+02 ]
>> Gradient at Termination: 169.85656457663794
>> ABNORMAL:
>> Estimation took: 02 minutes and 19 seconds
[12]:
source, target = meshes.values()
reconstructed = {}
for registration_dir in registration_dirs:
reconstructed[registration_dir.name] = PvSurface(
plddmm.io.load_deterministic_atlas_reconstruction(registration_dir, as_pv=True)
)
reconstructed["source"] = source
[13]:
{
name: varifold_metric.dist(target, reconstructed_)
for name, reconstructed_ in reconstructed.items()
}
[13]:
{'landmark': np.float64(15.674591642516685),
'landmark_svf': np.float64(16.995952344389686),
'varifold': np.float64(5.41578435415065),
'varifold_svf': np.float64(5.6515407334274705),
'source': np.float64(141.32513317492763)}
[14]:
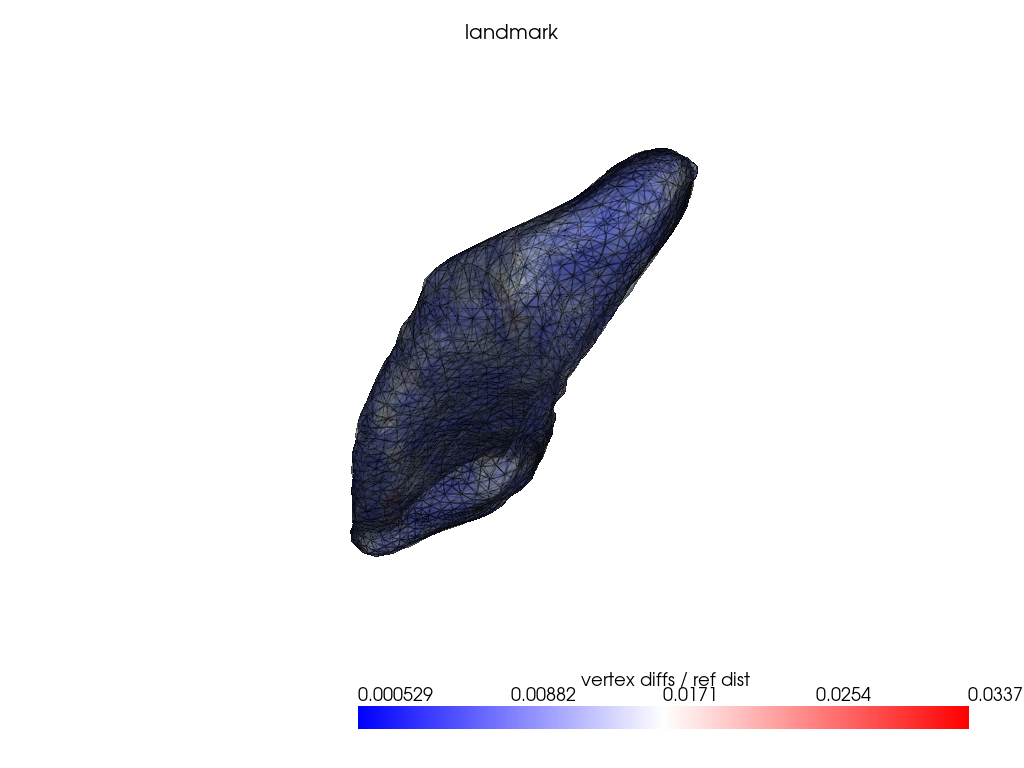
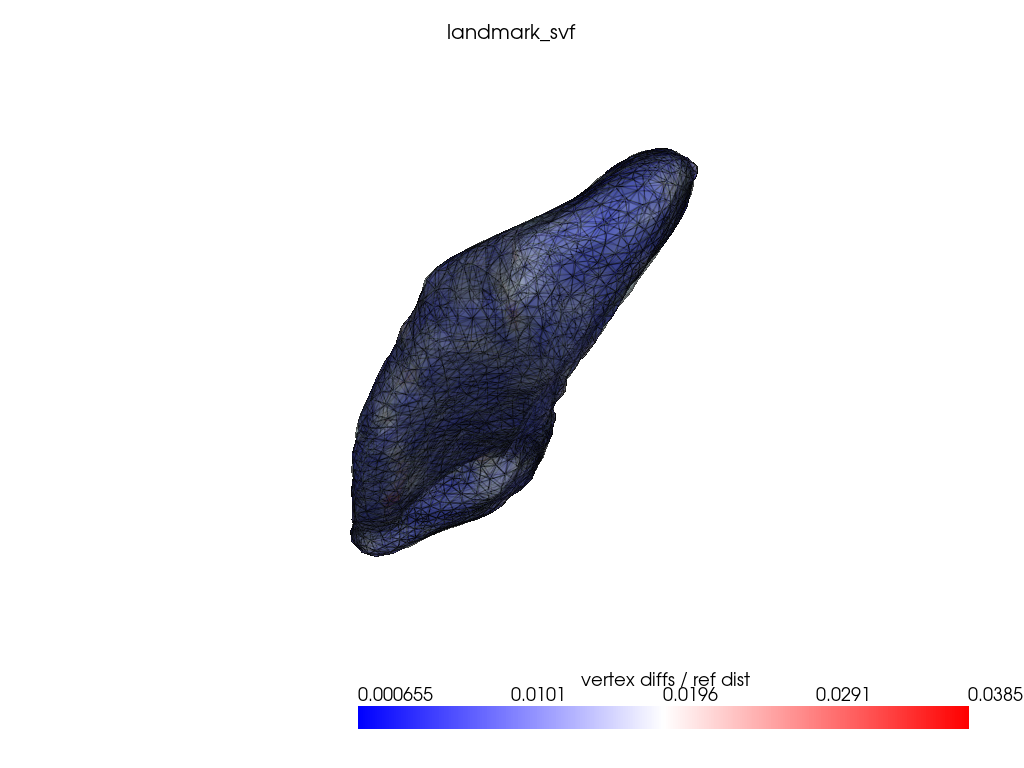
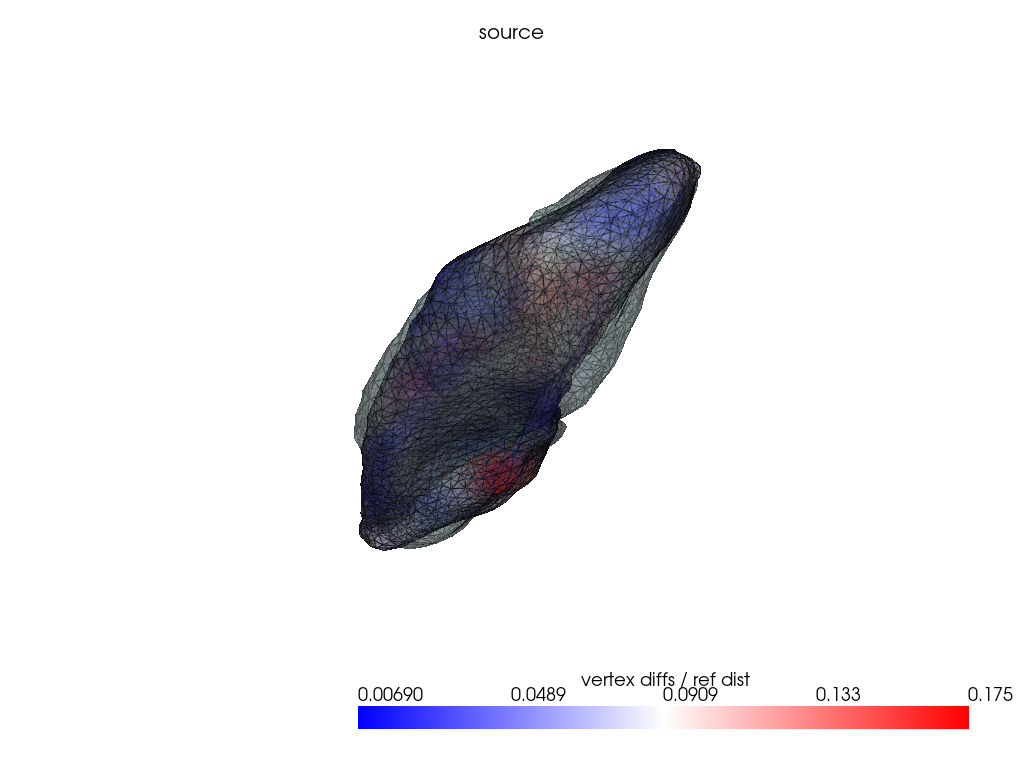
if VIZ > 0:
ref_dist = centroid2farthest_vertex([target])[0]
for name, reconstructed_ in reconstructed.items():
pl = RegisteredMeshesColoredPlotter()
pl.add_meshes(
target.as_pv(),
reconstructed_.as_pv(),
ref_dist=ref_dist,
show_edges=True,
opacity=0.8,
name="vertex diffs / ref dist",
)
pl.add_mesh(reconstructed_.as_pv(), show_edges=True, opacity=0.5)
pl.add_title(name, font_size=8.0)
pl.show()


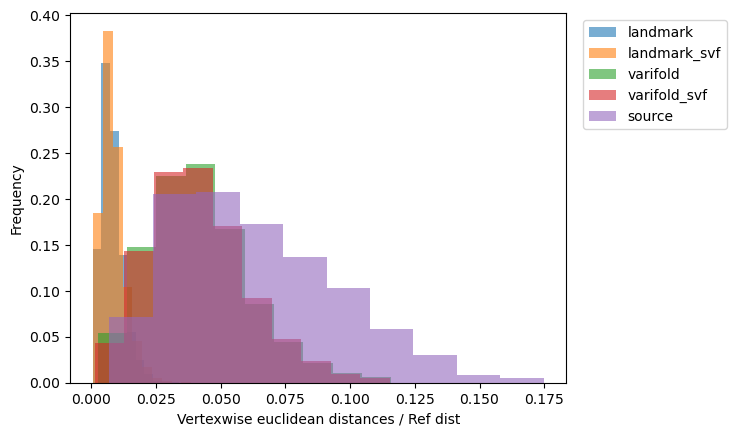
[15]:
_, ax = plt.subplots()
for name, reconstructed_ in reconstructed.items():
vals = vertexwise_euclidean(target, reconstructed_) / ref_dist
ax.hist(
vals,
weights=1 / len(vals) * np.ones_like(vals),
label=name,
alpha=0.6,
)
ax.set_xlabel("Vertexwise euclidean distances / Ref dist")
ax.set_ylabel("Frequency")
ax.legend(bbox_to_anchor=(1.02, 1), loc="upper left");