LDDMM: influence of regularisation#
[1]:
import shutil
from pathlib import Path
import numpy as np
import pyvista as pv
from matplotlib import pyplot as plt
import polpo.lddmm as plddmm
from polpo.mesh.qoi import centroid2farthest_vertex, vertexwise_euclidean
from polpo.mesh.surface import PvSurface
from polpo.mesh.varifold.tuning import SigmaFromLengths
from polpo.plot.pyvista import RegisteredMeshesColoredPlotter
from polpo.preprocessing.load.pregnancy.deformetrica import get_two_random_meshes
[KeOps] Warning : CUDA was detected, but driver API could not be initialized. Switching to CPU only.
[2]:
RECOMPUTE = False
STATIC_VIZ = True
VIZ = 1
if STATIC_VIZ:
pv.set_jupyter_backend("static")
[3]:
STRUCT_NAME = "L_Hipp"
OUTPUTS_DIR = Path.home() / ".polpo/results" / f"lddmm_regularisation_{STRUCT_NAME}"
REGISTRATION_DIR = OUTPUTS_DIR / "registration"
if OUTPUTS_DIR.exists() and RECOMPUTE:
shutil.rmtree(OUTPUTS_DIR)
[4]:
meshes = get_two_random_meshes(
OUTPUTS_DIR,
mesh_names=("source", "target"),
target_reduction=0,
)
[5]:
if VIZ > 1:
pl = pv.Plotter(border=False)
for mesh in meshes:
pl.add_mesh(mesh.as_pv(), show_edges=True, opacity=0.6)
pl.show()
We select the varifold kernel using characteristic lengths.
[6]:
sigma_search = SigmaFromLengths(
ratio_charlen_mesh=2.0,
ratio_charlen=0.25,
)
sigma_search.fit(meshes.values())
metric = sigma_search.optimal_metric_
sigma_search.sigma_
[6]:
np.float64(6.270582796758067)
Following LDDMM: how to register a mesh to a template?.
[7]:
mesh_filenames = list(meshes.keys())
def _registration_dir(regularisation):
return REGISTRATION_DIR / f"{str(regularisation).replace(".", "-")}"
regularisation = [0.01, 0.1, 1.0, 10.0]
registration_kwargs = dict(
kernel_width=4.0,
max_iter=2000,
freeze_control_points=False,
metric="varifold",
tol=1e-16,
attachment_kernel_width=sigma_search.sigma_,
)
for regularisation_ in regularisation:
registration_dir = _registration_dir(regularisation_)
if not registration_dir.exists():
plddmm.registration.estimate_registration(
mesh_filenames[0],
mesh_filenames[1],
output_dir=registration_dir,
regularisation=regularisation_,
**registration_kwargs,
)
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_regularisation_L_Hipp/registration/0-01/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -6.173E+05 [ attachment = -6.172E+05 ; regularity = -1.154E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -2.979E+05 [ attachment = -2.978E+05 ; regularity = -1.323E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -1.689E+05 [ attachment = -1.688E+05 ; regularity = -1.605E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -1.233E+05 [ attachment = -1.231E+05 ; regularity = -1.717E+02 ]
------------------------------------- Iteration: 100 -------------------------------------
>> Log-likelihood = -1.008E+05 [ attachment = -1.006E+05 ; regularity = -1.853E+02 ]
>> Gradient at Termination: 7111575109.681244
>> ABNORMAL:
>> Estimation took: 01 minutes and 51 seconds
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_regularisation_L_Hipp/registration/0-1/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -6.372E+03 [ attachment = -6.256E+03 ; regularity = -1.154E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -3.048E+03 [ attachment = -2.916E+03 ; regularity = -1.322E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -1.808E+03 [ attachment = -1.650E+03 ; regularity = -1.583E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -1.388E+03 [ attachment = -1.219E+03 ; regularity = -1.689E+02 ]
>> Gradient at Termination: 815750.447617195
>> ABNORMAL:
>> Estimation took: 01 minutes and 35 seconds
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_regularisation_L_Hipp/registration/1-0/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -1.673E+02 [ attachment = -6.219E+01 ; regularity = -1.051E+02 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -1.364E+02 [ attachment = -3.381E+01 ; regularity = -1.025E+02 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -1.268E+02 [ attachment = -2.512E+01 ; regularity = -1.017E+02 ]
------------------------------------- Iteration: 80 -------------------------------------
>> Log-likelihood = -1.229E+02 [ attachment = -2.331E+01 ; regularity = -9.959E+01 ]
------------------------------------- Iteration: 100 -------------------------------------
>> Log-likelihood = -1.206E+02 [ attachment = -2.091E+01 ; regularity = -9.969E+01 ]
>> Gradient at Termination: 71.2899148851723
>> ABNORMAL:
>> Estimation took: 01 minutes and 45 seconds
Logger has been set to: INFO
>> No initial CP spacing given: using diffeo kernel width of 4.0
OMP_NUM_THREADS was not found in environment variables. An automatic value will be set.
OMP_NUM_THREADS will be set to 10
context has already been set
>> No specified state-file. By default, Deformetrica state will by saved in file: /home/luisfpereira/.polpo/results/lddmm_regularisation_L_Hipp/registration/10-0/deformetrica-state.p.
>> Using a Sobolev gradient for the template data with the ScipyLBFGS estimator memory length being larger than 1. Beware: that can be tricky.
>> Set of 420 control points defined.
>> Momenta initialized to zero, for 1 subjects.
>> Started estimator: ScipyOptimize
>> Scipy optimization method: L-BFGS-B
------------------------------------- Iteration: 1 -------------------------------------
------------------------------------- Iteration: 20 -------------------------------------
>> Log-likelihood = -4.866E+01 [ attachment = -1.862E+01 ; regularity = -3.004E+01 ]
------------------------------------- Iteration: 40 -------------------------------------
>> Log-likelihood = -4.816E+01 [ attachment = -1.814E+01 ; regularity = -3.001E+01 ]
------------------------------------- Iteration: 60 -------------------------------------
>> Log-likelihood = -4.798E+01 [ attachment = -1.801E+01 ; regularity = -2.996E+01 ]
>> Gradient at Termination: 0.04452510888167644
>> ABNORMAL:
>> Estimation took: 01 minutes and 16 seconds
[8]:
reconstructed = {
regularisation_: PvSurface(
plddmm.io.load_deterministic_atlas_reconstruction(
_registration_dir(regularisation_), as_pv=True
)
)
for regularisation_ in regularisation
}
[9]:
_, target = meshes.values()
{
key: metric.dist(target, reconstructed_)
for key, reconstructed_ in reconstructed.items()
}
[9]:
{0.01: np.float64(2.8811387646548203),
0.1: np.float64(3.2341550700458614),
1.0: np.float64(4.474438161723856),
10.0: np.float64(42.38803313980611)}
[10]:
if VIZ > 0:
ref_dist = centroid2farthest_vertex([target])[0]
for name, reconstructed_ in reconstructed.items():
pl = RegisteredMeshesColoredPlotter()
pl.add_meshes(
target.as_pv(),
reconstructed_.as_pv(),
ref_dist=ref_dist,
show_edges=True,
opacity=0.8,
name="vertex diffs / ref dist",
)
pl.add_title(str(name), font_size=8.0)
pl.show()

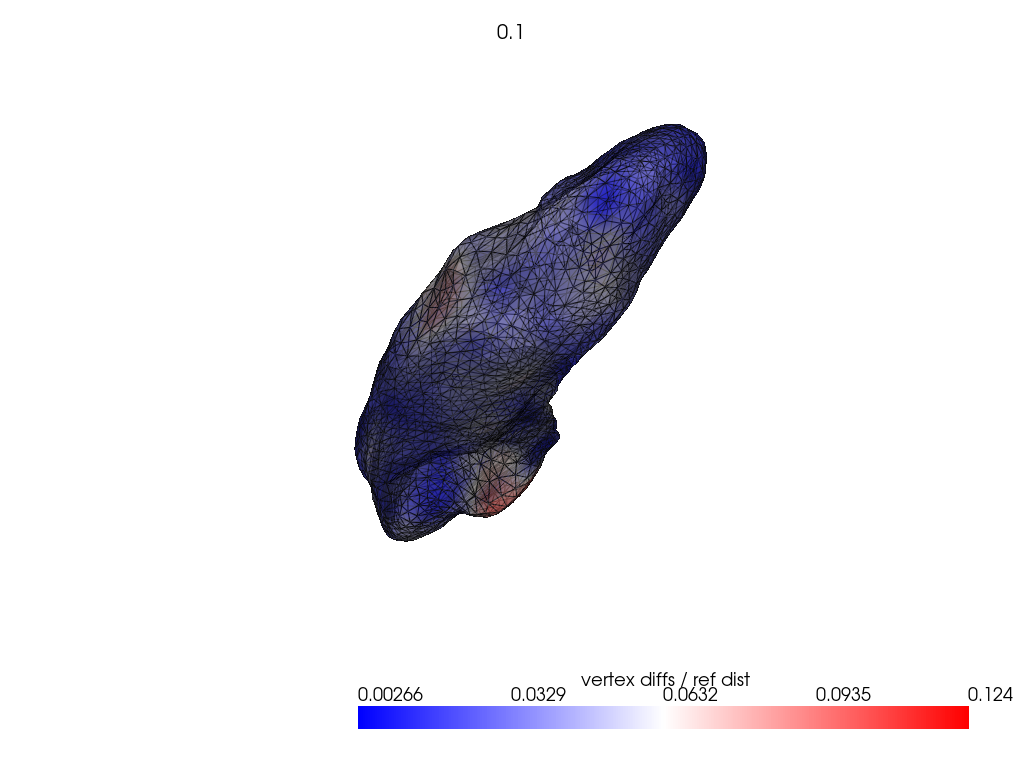
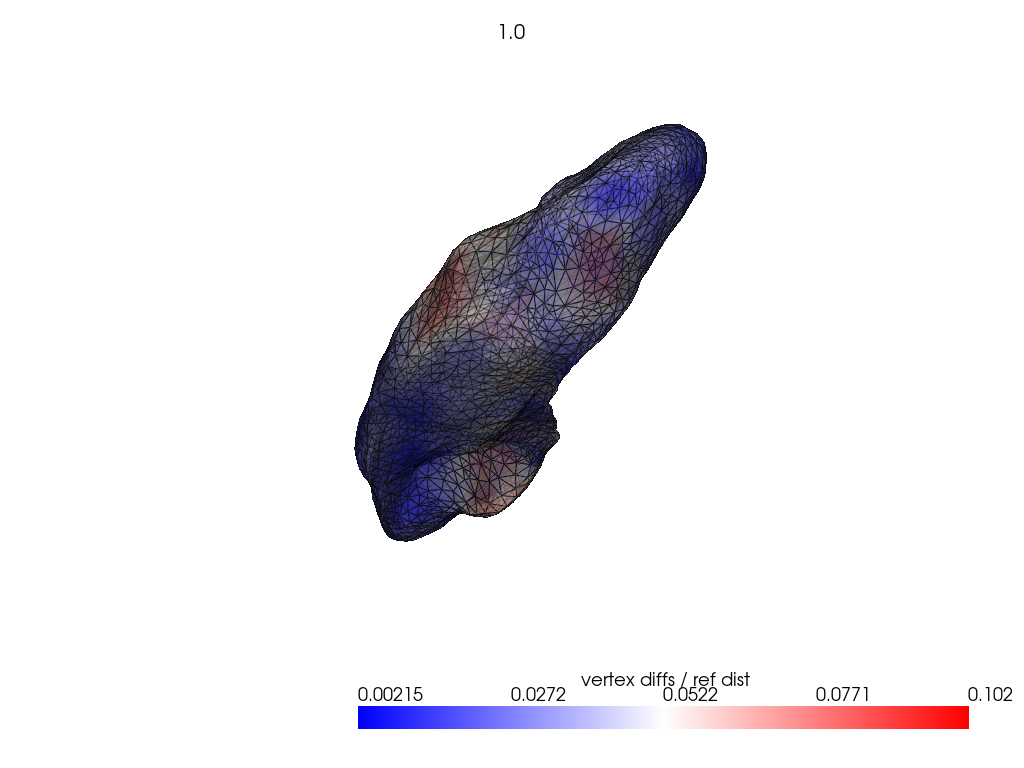
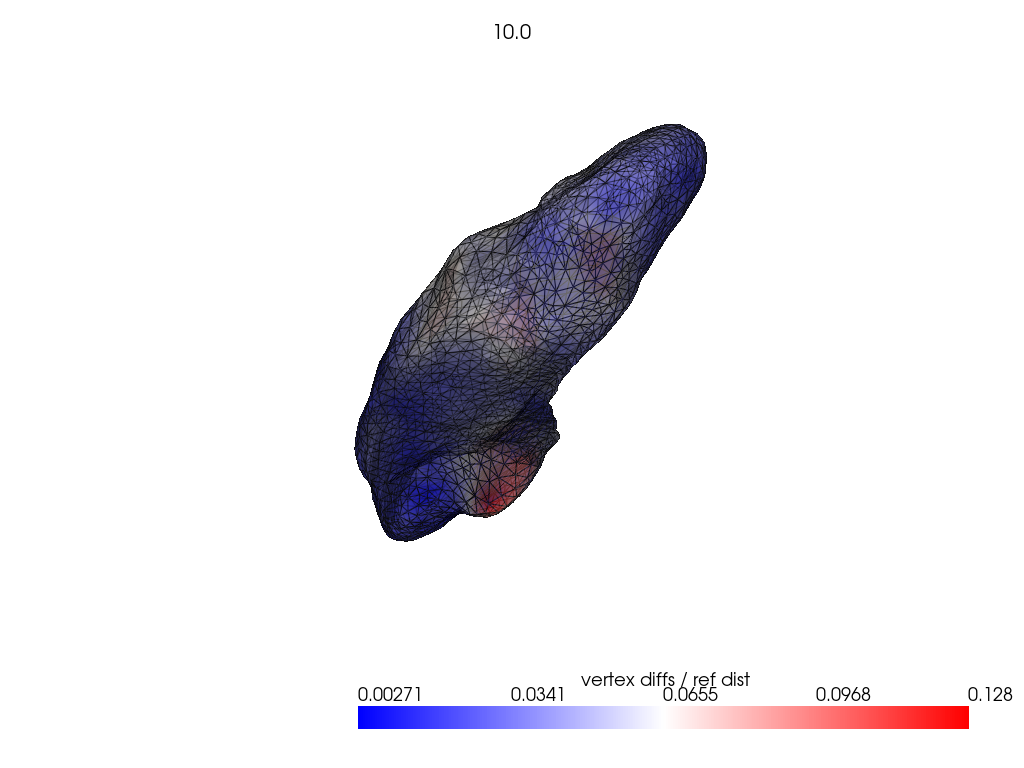
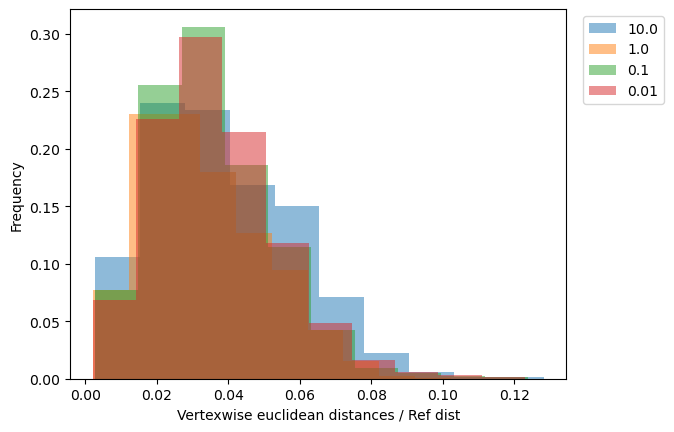
[11]:
_, ax = plt.subplots()
for name, reconstructed_ in reversed(reconstructed.items()):
vals = vertexwise_euclidean(target, reconstructed_) / ref_dist
ax.hist(
vals,
weights=1 / len(vals) * np.ones_like(vals),
label=name,
alpha=0.5,
)
ax.set_xlabel("Vertexwise euclidean distances / Ref dist")
ax.set_ylabel("Frequency")
ax.legend(bbox_to_anchor=(1.02, 1), loc="upper left");