Within dataset varifold distances#
Here we use a varifold metric to compute pairwise distances between meshes in a dataset.
[1]:
import numpy as np
from matplotlib import pyplot as plt
import polpo.preprocessing.dict as ppdict
from polpo.mesh.surface import PvSurface
from polpo.mesh.varifold.tuning import SigmaFromLengths
from polpo.preprocessing.load.pregnancy.jacobs import MeshLoader
[KeOps] Warning : CUDA was detected, but driver API could not be initialized. Switching to CPU only.
[2]:
mesh_loader = (
MeshLoader(
subject_subset=["01"],
struct_subset=["L_Hipp"],
derivative="enigma",
as_mesh=True,
)
+ ppdict.ExtractUniqueKey(nested=True)
+ ppdict.DictMap(PvSurface)
)
meshes = mesh_loader()
We select the varifold kernel using characteristic lengths.
[3]:
sigma_search = SigmaFromLengths(
ratio_charlen_mesh=2.0,
ratio_charlen=0.25,
)
sigma_search.fit([meshes[1]])
metric = sigma_search.optimal_metric_
sigma_search.sigma_
[3]:
np.float64(5.548036739288476)
[4]:
dists = []
for mesh in meshes.values():
dists_ = []
dists.append(dists_)
for cmp_mesh in meshes.values():
dists_.append(metric.dist(mesh, cmp_mesh))
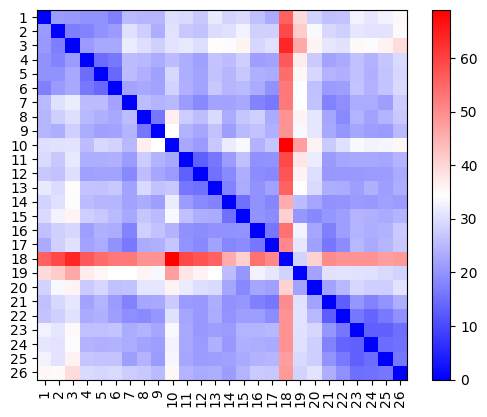
[36]:
fig, ax = plt.subplots()
im = ax.imshow(dists, cmap="bwr")
plt.colorbar(im)
ax.set_xticks(range(len(meshes)))
ax.set_xticklabels(meshes.keys(), rotation=90)
ax.set_yticks(range(len(meshes)))
ax.set_yticklabels(meshes.keys());
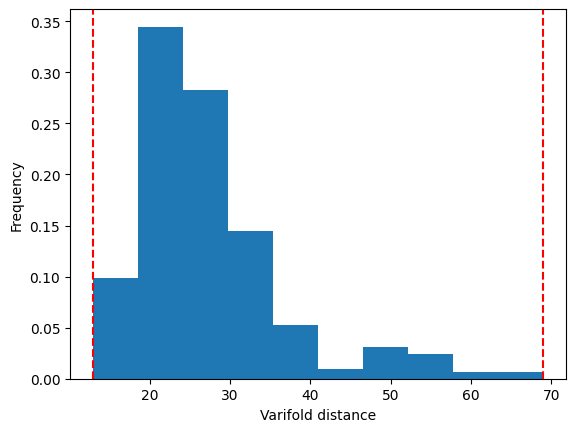
[34]:
dists_ = np.array(dists)[np.triu_indices(len(dists), k=1)]
_, ax = plt.subplots()
ax.hist(
dists_,
weights=1 / len(dists_) * np.ones_like(dists_),
)
ax.set_xlabel("Varifold distance")
ax.set_ylabel("Frequency")
min_dist, max_dist = np.amin(dists_), np.amax(dists_)
ax.axvline(min_dist, color="red", linestyle="--")
ax.axvline(max_dist, color="red", linestyle="--")
min_dist, max_dist
[34]:
(np.float64(12.870735508410826), np.float64(69.00952156307977))