Impact of decimation in the automatic selection of the varifold kernel width using a decimation-based approach#
Before, we showed how a varifold kernel can be selected automatically using a decimation-based approach.
Here, we show some of the drawbacks of the method.
[1]:
import numpy as np
from matplotlib import pyplot as plt
import polpo.preprocessing.dict as ppdict
from polpo.mesh.surface import PvSurface
from polpo.mesh.varifold.tuning import SigmaBisecSearch
from polpo.preprocessing import BranchingPipeline, IdentityStep
from polpo.preprocessing.load.pregnancy.jacobs import MeshLoader
from polpo.preprocessing.mesh.decimation import PvDecimate
[KeOps] Warning : CUDA was detected, but driver API could not be initialized. Switching to CPU only.
[2]:
session_subset = np.random.choice(np.arange(1, 27))
mesh_loader = (
MeshLoader(
subject_subset=["01"],
struct_subset=["L_Hipp"],
session_subset=[session_subset],
derivative="enigma",
as_mesh=True,
)
+ ppdict.ExtractUniqueKey(nested=True)
+ PvSurface
)
raw_mesh = mesh_loader()
session_subset
[2]:
np.int64(21)
[3]:
decimation_targets = [0.8, 0.5, 0.2, 0]
multi_decimation_pipe = BranchingPipeline(
[
(
PvDecimate(target_reduction=target_reduction, volume_preservation=True)
+ PvSurface
if target_reduction
else IdentityStep()
)
for target_reduction in decimation_targets
],
merger=lambda data: dict(zip(decimation_targets, data)),
)
meshes = multi_decimation_pipe(raw_mesh)
[mesh.n_points for mesh in meshes.values()]
[3]:
[502, 1252, 2002, 2502]
How does decimation affect mass?
[4]:
[np.sum(mesh.face_areas) for mesh in meshes.values()]
[4]:
[np.float64(1734.77822369891),
np.float64(1734.7171298008561),
np.float64(1734.7350274907199),
np.float64(1734.7065791328196)]
[5]:
[np.sum(mesh.volume) for mesh in meshes.values()]
[5]:
[np.float64(3478.998321966246),
np.float64(3489.383964996294),
np.float64(3490.5027858115213),
np.float64(3490.617830330979)]
[6]:
ref_value = 0.1
grid_search = SigmaBisecSearch(
ref_value=ref_value,
decimator=True,
)
[7]:
sigmas = {}
sdists = {}
for target_red, mesh in meshes.items():
grid_search.fit([mesh])
sigmas[target_red] = grid_search.grid_.copy()
sdists[target_red] = grid_search.sdists_.copy()
/home/luisfpereira/Repos/github/polpo/polpo/mesh/varifold/tuning.py:255: UserWarning: No convergence. Upper interval bound is too strict.
warnings.warn("No convergence. Upper interval bound is too strict.")
[8]:
_, ax = plt.subplots()
for target_red in meshes:
ax.scatter(sigmas[target_red], sdists[target_red], label=target_red)
x_min, x_max = ax.get_xlim()
ax.hlines(y=ref_value, xmin=x_min, xmax=x_max, linestyle="--")
ax.set_xlabel(r"$\sigma$")
ax.set_ylabel(r"$\Sigma d_i$")
ax.legend(title="Reduc", bbox_to_anchor=(1.02, 1), loc="upper left")
ax.set_ylim([0, 0.4])
[8]:
(0.0, 0.4)
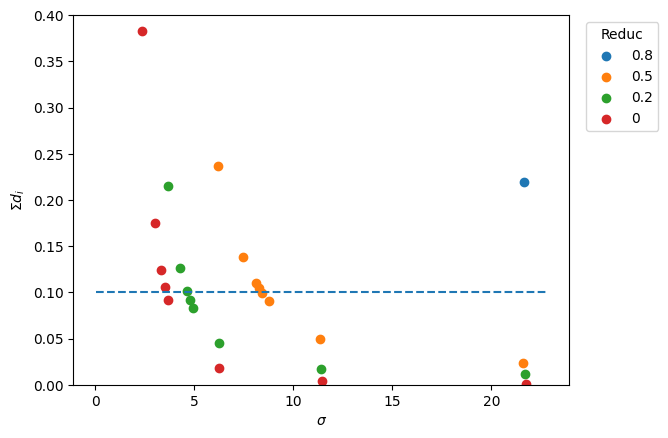
The culprit.
[9]:
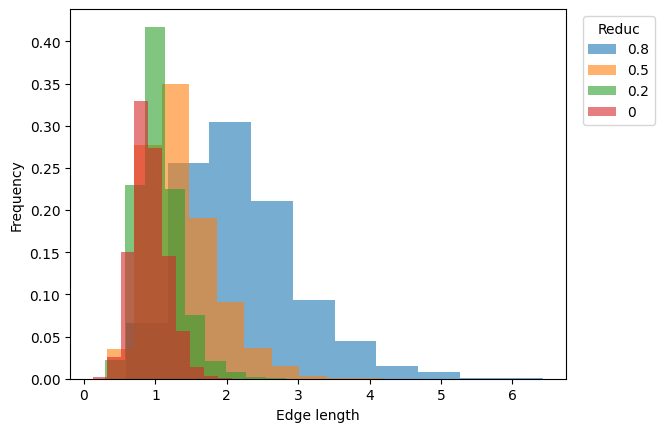
[np.amax(mesh.edge_lengths) for mesh in meshes.values()]
[9]:
[np.float32(6.4342594),
np.float32(4.1676106),
np.float32(3.1141806),
np.float64(2.0772219039275552)]
[10]:
_, ax = plt.subplots()
for target_red, mesh in meshes.items():
edge_lengths = mesh.edge_lengths
ax.hist(
edge_lengths,
label=target_red,
alpha=0.6,
weights=1 / edge_lengths.size * np.ones_like(edge_lengths),
)
ax.set_xlabel("Edge length")
ax.set_ylabel("Frequency")
ax.legend(title="Reduc", bbox_to_anchor=(1.02, 1), loc="upper left");