Notebook source code: notebooks/02_methods_estimate_manifold_dimension.ipynb
Estimate Neural Dimensions#
This notebook explains how to use the module dimension to estimate the dimension (extrinsic or intrinsic) of the neural manifold embedded in neural state space \(\mathbb{R}^N\).
In particular, this notebook tests several existing methods of dimension estimation on the synthetic manifolds from the module datasets.synthetic to evaluate their performance.
Set-up + Imports#
In [1]:
import setup
setup.main()
%load_ext autoreload
%autoreload 2
%load_ext jupyter_black
import os
import matplotlib.pyplot as plt
import numpy as np
import skdim
import neurometry.datasets.synthetic as synthetic
from neurometry.dimension.dimension import (
plot_dimension_experiments,
skdim_dimension_estimation,
)
import torch
os.environ["GEOMSTATS_BACKEND"] = "pytorch"
import geomstats.backend as gs
Working directory: /home/facosta/neurometry/neurometry
Directory added to path: /home/facosta/neurometry
Directory added to path: /home/facosta/neurometry/neurometry
INFO:root:Using pytorch backend
INFO:numexpr.utils:Note: NumExpr detected 32 cores but "NUMEXPR_MAX_THREADS" not set, so enforcing safe limit of 8.
INFO:numexpr.utils:NumExpr defaulting to 8 threads.
Show Methods Implemented in \(\texttt{scikit-dimension}\)#
In [153]:
all_methods = [method for method in dir(skdim.id) if not method.startswith("_")]
print(all_methods)
['CorrInt', 'DANCo', 'ESS', 'FisherS', 'KNN', 'MADA', 'MLE', 'MOM', 'MiND_ML', 'TLE', 'TwoNN', 'lPCA']
Define Parameters for Experiments#
In [150]:
num_points = 2000
num_neurons = 40
max_id_dim = 20
dimensions = [_ for _ in range(1, max_id_dim + 1)]
methods = ["MLE", "lPCA", "TwoNN", "CorrInt"]
poisson_multiplier = 1
num_trials = 20
ref_frequency = 50
Run Experiments for Hyperspheres with \(\texttt{scikit-dimension}\)#
In [151]:
manifold_type = "hypersphere"
id_estimates_hyperspheres, noise_level = skdim_dimension_estimation(
methods,
dimensions,
manifold_type,
num_trials,
num_points,
num_neurons,
poisson_multiplier,
ref_frequency,
)
plot_dimension_experiments(
id_estimates_hyperspheres, dimensions, max_id_dim, manifold_type, noise_level
)
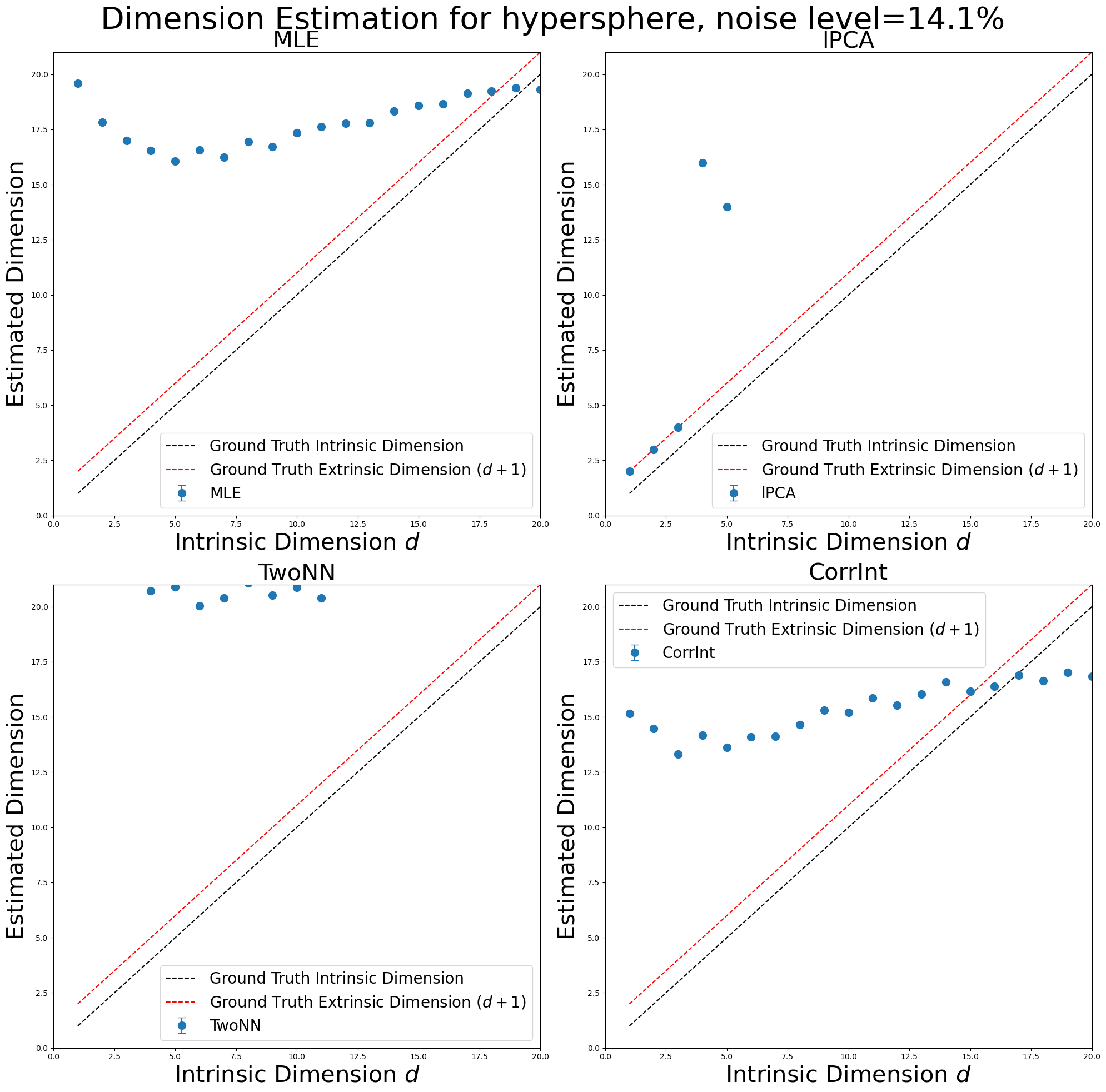
Run Experiments for Hypertori with \(\texttt{scikit-dimension}\)#
In [152]:
manifold_type = "hypertorus"
id_estimates_hypertori, noise_level = skdim_dimension_estimation(
methods,
dimensions,
manifold_type,
num_trials,
num_points,
num_neurons,
poisson_multiplier,
ref_frequency,
)
plot_dimension_experiments(
id_estimates_hypertori, dimensions, max_id_dim, manifold_type, noise_level
)
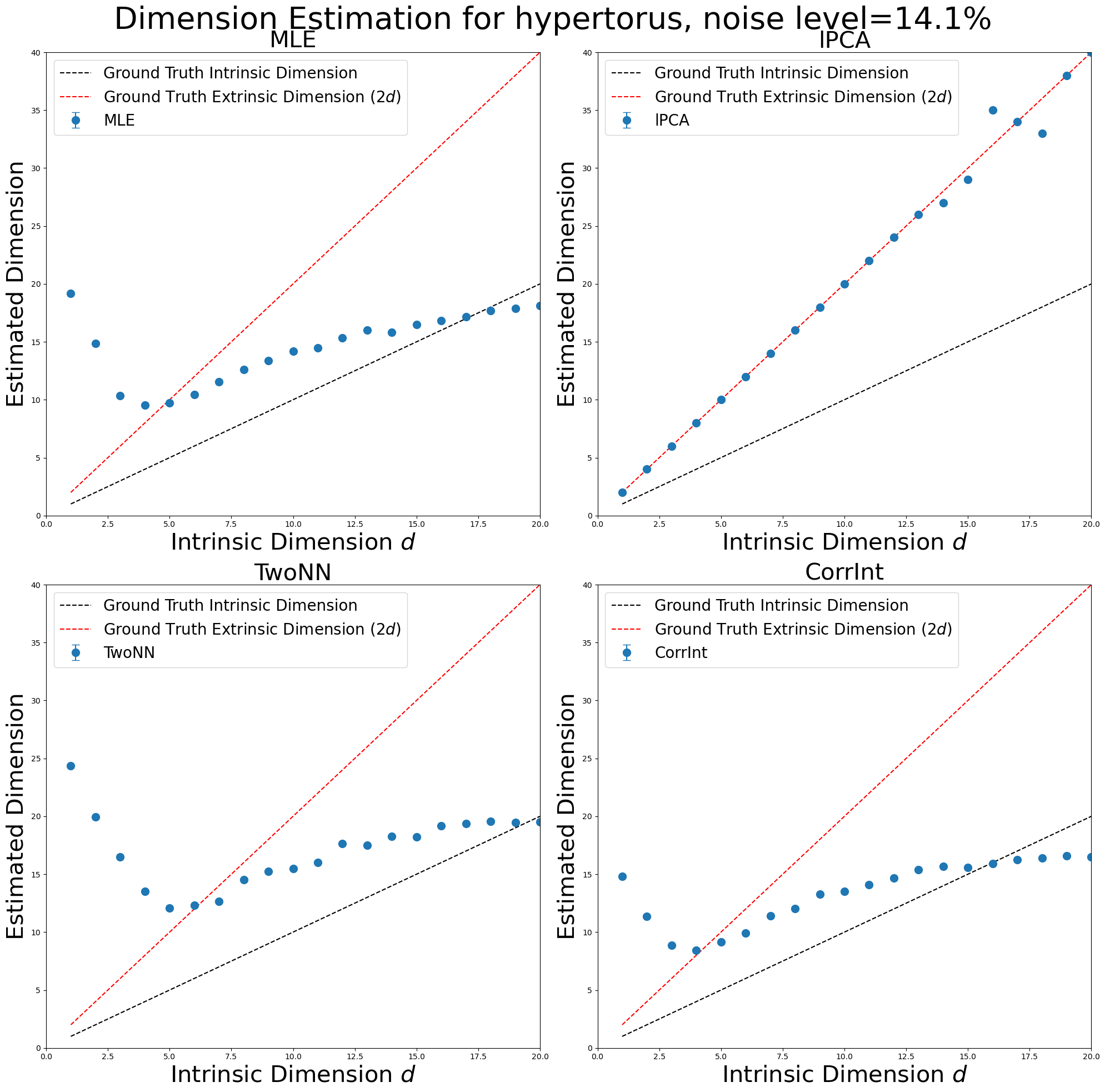
Try Single Experiment#
In [8]:
num_points = 2000
num_neurons = 40
poisson_multiplier = 1
num_trials = 50
ref_frequency = 50
manifold_type = "hypertorus"
point_generator = getattr(synthetic, manifold_type)
points = point_generator(intrinsic_dim=1, num_points=num_points)
neural_manifold, _ = synthetic.synthetic_neural_manifold(
points,
num_neurons,
"sigmoid",
poisson_multiplier,
ref_frequency,
scales=gs.ones(num_neurons),
)
In [9]:
neural_manifold.shape
Out [9]:
torch.Size([2000, 40])
In [217]:
method_name = "KNN"
method = getattr(skdim.id, method_name)()
estimates = np.zeros(num_trials)
for trial_idx in range(num_trials):
method.fit(neural_manifold)
estimates[trial_idx] = np.mean(method.dimension_)
print(estimates)
[ 2. 2. 3. 5. 2. 2. 4. 2. 2. 2. 2. 17. 4. 2. 40. 2. 2. 2.
5. 3. 2. 2. 2. 3. 40. 2. 2. 11. 10. 5. 40. 2. 4. 4. 38. 3.
3. 3. 1. 3. 2. 40. 2. 3. 2. 7. 2. 9. 2. 2.]
try: find median/mode of KNN algorithm
In [218]:
torch.median(torch.tensor(estimates))
Out [218]:
tensor(2.)
try: first PCA data down to minimal embedding dim, then nonlinear dim est
PLS approach#
In [48]:
import numpy as np
from sklearn.cross_decomposition import PLSRegression
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
from sklearn.multioutput import MultiOutputRegressor
def evaluate_pls_with_different_K(X, Y, K_values):
"""
Evaluate PLS Regression followed by Multi-Output Regression for different numbers of components (K).
Parameters:
- X: Neural activity data (predictors)
- Y: Continuous 2D outcomes
- K_values: A list of integers representing different numbers of PLS components to evaluate
Returns:
- A list of R^2 scores corresponding to each K-value
"""
r2_scores = []
projected_X = []
# Split data into training and test sets
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
for K in K_values:
# Initialize and fit PLS Regression
pls = PLSRegression(n_components=K)
pls.fit(X_train, Y_train)
# Project both training and test data using the PLS model
X_train_pls = pls.transform(X_train)
X_test_pls = pls.transform(X_test)
projected_X.append(pls.inverse_transform(X_test_pls))
# Fit the Multi-Output Regression model on the reduced data
multi_output_reg = MultiOutputRegressor(LinearRegression()).fit(
X_train_pls, Y_train
)
# Predict and evaluate using R^2 score
Y_pred = multi_output_reg.predict(X_test_pls)
score = r2_score(
Y_test, Y_pred, multioutput="uniform_average"
) # Average R^2 score across all outputs
r2_scores.append(score)
return r2_scores, projected_X
In [49]:
from sklearn.decomposition import PCA
def evaluate_PCA_with_different_K(X, Y, K_values):
"""
Evaluate PCA for different numbers of components (K).
Parameters:
- X: Data to perform PCA on
- K_values: A list of integers representing different numbers of PCA components to evaluate
Returns:
- A list of R^2 scores corresponding to each K-value
"""
r2_scores = []
projected_X = []
# Split data into training and test sets
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
for K in K_values:
# Initialize and fit PCA
pca = PCA(n_components=K)
pca.fit(X_train)
# Project both training and test data using the PCA model
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
projected_X.append(pca.inverse_transform(X_test_pca))
# Fit the Multi-Output Regression model on the reduced data
multi_output_reg = MultiOutputRegressor(LinearRegression()).fit(
X_train_pca, Y_train
)
# Predict and evaluate using R^2 score
Y_pred = multi_output_reg.predict(X_test_pca)
score = r2_score(
Y_test, Y_pred, multioutput="uniform_average"
) # Average R^2 score across all outputs
r2_scores.append(score)
return r2_scores, projected_X
In [65]:
import numpy as np
from sklearn.model_selection import KFold
def evaluate_PCA_with_different_K_cv(X, Y, K_values, n_splits=5):
"""
Evaluate PCA for different numbers of components (K) using cross-validation.
Parameters:
- X: Data to perform PCA on
- Y: Continuous 2D outcomes
- K_values: A list of integers representing different numbers of PCA components to evaluate
- n_splits: Number of folds in K-Fold cross-validation
Returns:
- A dictionary of R^2 scores for each K-value, each containing a list of scores for each fold
"""
cv_scores = {
K: [] for K in K_values
} # Initialize dictionary to store scores for each K
kf = KFold(n_splits=n_splits, shuffle=True, random_state=42)
for K in K_values:
# Loop through each fold defined by KFold
for train_index, test_index in kf.split(X):
# Split data into training and test sets for this fold
X_train, X_test = X[train_index], X[test_index]
Y_train, Y_test = Y[train_index], Y[test_index]
# Initialize and fit PCA
pca = PCA(n_components=K)
pca.fit(X_train)
# Project both training and test data using the PCA model
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
# Fit the Multi-Output Regression model on the reduced data
multi_output_reg = MultiOutputRegressor(LinearRegression()).fit(
X_train_pca, Y_train
)
# Predict and evaluate using R^2 score
Y_pred = multi_output_reg.predict(X_test_pca)
score = r2_score(Y_test, Y_pred, multioutput="uniform_average")
cv_scores[K].append(score)
return cv_scores
In [66]:
import numpy as np
def evaluate_pls_with_different_K_cv(X, Y, K_values, n_splits=5):
"""
Evaluate PLS Regression followed by Multi-Output Regression for different numbers of components (K)
using cross-validation.
Parameters:
- X: Neural activity data (predictors)
- Y: Continuous 2D outcomes
- K_values: A list of integers representing different numbers of PLS components to evaluate
- n_splits: Number of folds in K-Fold cross-validation
Returns:
- A dictionary of R^2 scores for each K-value, each containing a list of scores for each fold
"""
cv_scores = {
K: [] for K in K_values
} # Initialize dictionary to store scores for each K
kf = KFold(n_splits=n_splits, shuffle=True, random_state=42)
for K in K_values:
# Loop through each fold defined by KFold
for train_index, test_index in kf.split(X):
# Split data into training and test sets for this fold
X_train, X_test = X[train_index], X[test_index]
Y_train, Y_test = Y[train_index], Y[test_index]
# Initialize and fit PLS Regression
pls = PLSRegression(n_components=K)
pls.fit(X_train, Y_train)
# Project both training and test data using the PLS model
X_train_pls = pls.transform(X_train)
X_test_pls = pls.transform(X_test)
# Fit the Multi-Output Regression model on the reduced data
multi_output_reg = MultiOutputRegressor(LinearRegression()).fit(
X_train_pls, Y_train
)
# Predict and evaluate using R^2 score
Y_pred = multi_output_reg.predict(X_test_pls)
score = r2_score(
Y_test, Y_pred, multioutput="uniform_average"
) # Average R^2 score across all outputs
cv_scores[K].append(score)
return cv_scores
In [90]:
import neurometry.datasets.synthetic as synthetic
N = 20
task_points = synthetic.hypertorus(2, 500)
noisy_points, manifold_points = synthetic.synthetic_neural_manifold(
points=task_points,
encoding_dim=N,
nonlinearity="sigmoid",
poisson_multiplier=1,
scales=gs.array(np.random.uniform(1, 3, N)),
)
noise level: 7.07%
In [91]:
import numpy as np
import torch
def add_noise_dimensions(data, n_dims):
"""
Adds a column of random noise to the input data.
Parameters:
- data: A numpy array of shape (num_points, dim), where `num_points` is the number of data points
and `dim` is the dimensionality of each data point.
- noise_scale: The standard deviation of the Gaussian noise to be added. Default is 1.0.
Returns:
- A numpy array of shape (num_points, dim+1), where the last column is filled with random noise.
"""
num_points, dim = data.shape
noise = np.random.normal(
loc=torch.mean(data), scale=1.5 * torch.std(data), size=(num_points, n_dims)
)
# noise = np.random.uniform(
# low=torch.min(data), high=torch.max(data), size=(num_points, n_dims)
# )
noisy_data = np.hstack((data, noise))
return noisy_data
In [92]:
X = add_noise_dimensions(noisy_points, n_dims=N)
Y = task_points
print(noisy_points.shape)
print(X.shape)
plt.imshow(X.T, aspect="auto", interpolation="None");
torch.Size([500, 20])
(500, 40)
In [93]:
K_values = [i for i in range(1, N + 1)]
pls_r2_scores, pls_transformed_X = evaluate_pls_with_different_K(X, Y, K_values)
pca_r2_scores, pca_transformed_X = evaluate_PCA_with_different_K(X, Y, K_values)
plt.plot(K_values, pls_r2_scores, marker="o", label="PLS $R^2$ Score")
plt.plot(K_values, pca_r2_scores, marker="o", label="PCA $R^2$ Score")
# pls_cv_scores = evaluate_pls_with_different_K_cv(X, Y, K_values, n_splits=5)
# pca_cv_scores = evaluate_PCA_with_different_K_cv(X, Y, K_values, n_splits=5)
# plt.errorbar(
# K_values,
# np.mean(list(pls_cv_scores.values()), axis=1),
# yerr=np.std(list(pls_cv_scores.values()), axis=1),
# fmt="-o",
# label="PLS R^2 Score",
# )
# plt.errorbar(
# K_values,
# np.mean(list(pca_cv_scores.values()), axis=1),
# yerr=np.std(list(pca_cv_scores.values()), axis=1),
# fmt="-o",
# label="PCA R^2 Score",
# )
plt.xticks(K_values)
plt.xlabel("Number of PLS/PCA Components (K)")
plt.ylabel("$R^2$ Score of Position Prediction")
plt.title("Position Prediction Performance vs. Number of Components")
plt.legend(loc="center left", bbox_to_anchor=(1, 0.9));
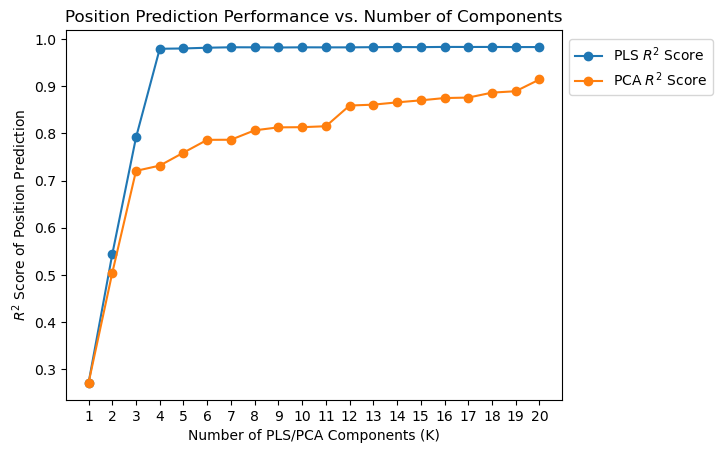
In [94]:
from gtda.homology import VietorisRipsPersistence
from gtda.plotting import plot_diagram
In [95]:
n_components = 4
X_pls = pls_transformed_X[n_components - 1]
X_pca = pca_transformed_X[n_components - 1]
vr_pers = VietorisRipsPersistence(homology_dimensions=(0, 1, 2), n_jobs=-1)
diagrams = vr_pers.fit_transform([X_pls, X_pca])
In [96]:
pls_pers_plot = plot_diagram(diagrams[0])
pls_pers_plot.update_layout(title="V-R Persist. Diag., PLS-projected data (dim=4)")
In [97]:
pca_pers_plot = plot_diagram(diagrams[1])
pca_pers_plot.update_layout(
title=f"V-R Persist. Diag., PCA-projected data (dim={n_components})"
)
In [77]:
from gtda.diagrams import BettiCurve, Scaler
def max_function(x):
return np.max(x)
scaler = Scaler(function=max_function)
diagrams_scaled = scaler.fit_transform(diagrams)
In [98]:
betticurves = BettiCurve(n_bins=50, n_jobs=-1)
betti_curves = betticurves.fit_transform(diagrams)
In [99]:
betticurves.plot(betti_curves, sample=0)
In [100]:
# n_components = 4
# X_pls = pls_transformed_X[n_components - 1]
# X_pca = pca_transformed_X[n_components - 1]
# from gtda.homology import WeakAlphaPersistence
# wa_pers = WeakAlphaPersistence(homology_dimensions=(0, 1, 2))
# diagrams_wa_pers = wa_pers.fit_transform([X_pca, X_pls])
---------------------------------------------------------------------------
_RemoteTraceback Traceback (most recent call last)
_RemoteTraceback:
"""
Traceback (most recent call last):
File "/home/facosta/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/externals/loky/process_executor.py", line 463, in _process_worker
r = call_item()
File "/home/facosta/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/externals/loky/process_executor.py", line 291, in __call__
return self.fn(*self.args, **self.kwargs)
File "/home/facosta/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py", line 589, in __call__
return [func(*args, **kwargs)
File "/home/facosta/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py", line 589, in <listcomp>
return [func(*args, **kwargs)
File "/home/facosta/miniconda3/envs/neurometry/lib/python3.9/site-packages/gtda/homology/simplicial.py", line 1110, in _weak_alpha_diagram
indptr, indices = Delaunay(X).vertex_neighbor_vertices
File "_qhull.pyx", line 1827, in scipy.spatial._qhull.Delaunay.__init__
File "_qhull.pyx", line 353, in scipy.spatial._qhull._Qhull.__init__
scipy.spatial._qhull.QhullError: QH6154 Qhull precision error: Initial simplex is flat (facet 1 is coplanar with the interior point)
While executing: | qhull d Qt Q12 Qx Qc Qz Qbb
Options selected for Qhull 2019.1.r 2019/06/21:
run-id 1336544124 delaunay Qtriangulate Q12-allow-wide Qxact-merge
Qcoplanar-keep Qz-infinity-point Qbbound-last _zero-centrum
Qinterior-keep Q3-no-merge-vertices-dim-high Pgood _max-width 2.8e+02
Error-roundoff 1.4e-11 _one-merge 1.2e-09 Visible-distance 8.6e-11
U-max-coplanar 8.6e-11 Width-outside 1.7e-10 _wide-facet 5.2e-10
_maxoutside 1.2e-09
precision problems (corrected unless 'Q0' or an error)
42 nearly singular or axis-parallel hyperplanes
7 zero divisors during gaussian elimination
The input to qhull appears to be less than 41 dimensional, or a
computation has overflowed.
Qhull could not construct a clearly convex simplex from points:
- p14(v42): 1.6e+02 68 1e+02 91 1.6e+02 1e+02 40 1.5e+02 1.2e+02 1e+02 1.3e+02 71 85 1.2e+02 40 69 90 64 1.3e+02 1.1e+02 1e+02 1.1e+02 95 1.1e+02 95 1.3e+02 96 85 78 92 1e+02 1e+02 1.1e+02 73 1.2e+02 45 97 68 98 78 23
- p13(v41): 2e+02 41 64 1.1e+02 1.6e+02 1.1e+02 1.3e+02 1.2e+02 1.5e+02 1.3e+02 2e+02 48 71 51 56 44 45 1.4e+02 1.2e+02 91 1.2e+02 1.2e+02 1e+02 60 91 1.2e+02 84 90 99 74 99 69 72 1.2e+02 1.1e+02 82 1e+02 91 1.1e+02 1e+02 71
- p12(v40): -3.9 1.4e+02 1.1e+02 22 40 1.3e+02 1.1e+02 24 1.3 32 77 1.1e+02 1.1e+02 1.7e+02 1.6e+02 1.8e+02 2e+02 47 1.4e+02 1.8e+02 98 1.2e+02 82 67 61 1.3e+02 1e+02 89 81 88 86 68 1e+02 1.1e+02 1e+02 74 85 1e+02 1.3e+02 93 76
- p11(v39): 1.9e+02 53 66 1.4e+02 1.5e+02 1e+02 1.3e+02 1.3e+02 1.6e+02 1.4e+02 1.8e+02 65 82 37 61 41 32 1.5e+02 95 64 1.2e+02 1.1e+02 1e+02 75 1e+02 1.1e+02 88 1e+02 1.1e+02 83 1e+02 86 78 1.2e+02 1e+02 1e+02 98 98 1.1e+02 1e+02 80
- p10(v38): 1.9e+02 54 1.2e+02 1.1e+02 1.8e+02 70 3.5 1.9e+02 1.7e+02 1.2e+02 1.2e+02 78 81 1e+02 19 42 50 65 1.2e+02 70 89 87 1.2e+02 1.8e+02 1.5e+02 74 93 1.1e+02 1.3e+02 1.3e+02 80 1.6e+02 1.1e+02 52 1.3e+02 1.1e+02 1.2e+02 77 79 76 1.2e+02
- p9(v37): 64 98 82 68 65 1.2e+02 1.8e+02 22 77 73 1.5e+02 89 86 72 1.6e+02 1.3e+02 1.1e+02 1.4e+02 1.1e+02 1.3e+02 1.1e+02 1.1e+02 1.1e+02 71 1e+02 66 84 1.1e+02 1.6e+02 1e+02 59 72 54 1.5e+02 1.1e+02 1.8e+02 1.1e+02 1.4e+02 1.3e+02 1.2e+02 1.1e+02
- p8(v36): 1e+02 79 97 54 1.1e+02 1.2e+02 1.2e+02 67 90 68 1.5e+02 74 77 99 1.2e+02 1.1e+02 1.1e+02 96 1.4e+02 1.4e+02 1e+02 1.2e+02 1.1e+02 95 1e+02 81 85 1e+02 1.4e+02 1.1e+02 62 86 68 1.1e+02 1.2e+02 1.4e+02 1.1e+02 1.2e+02 1.2e+02 1e+02 55
- p7(v35): 32 1.3e+02 1.2e+02 33 68 1.3e+02 73 60 18 45 81 1e+02 1e+02 1.8e+02 1.2e+02 1.6e+02 1.9e+02 32 1.4e+02 1.8e+02 99 1.2e+02 76 69 56 1.5e+02 1e+02 80 57 80 1e+02 71 1.2e+02 92 1e+02 34 80 83 1.2e+02 82 39
- p6(v34): 26 1.3e+02 86 44 46 1.4e+02 1.5e+02 16 13 54 1.2e+02 95 1e+02 1.4e+02 1.6e+02 1.6e+02 1.8e+02 86 1.3e+02 1.7e+02 1.1e+02 1.3e+02 76 20 42 1.5e+02 98 83 63 61 1e+02 36 93 1.3e+02 90 56 76 1e+02 1.4e+02 1e+02 59
- p4(v33): 1.2e+02 69 83 45 1.2e+02 1.4e+02 1.1e+02 69 74 68 1.8e+02 55 72 1.1e+02 98 1.1e+02 1.2e+02 83 1.5e+02 1.6e+02 1.1e+02 1.3e+02 96 49 67 1.3e+02 86 77 92 75 85 52 75 1.1e+02 1.2e+02 69 99 93 1.2e+02 99 32
- p3(v32): 1.4e+02 76 68 1.1e+02 1.1e+02 1.2e+02 1.5e+02 86 1.2e+02 1.2e+02 1.6e+02 75 88 59 98 77 71 1.5e+02 1e+02 94 1.2e+02 1.1e+02 97 54 87 1.2e+02 89 1e+02 1e+02 76 1e+02 68 77 1.3e+02 98 98 92 1.1e+02 1.2e+02 1.1e+02 42
- p2(v31): 2.1e+02 53 77 1.3e+02 1.8e+02 1e+02 56 1.8e+02 1.4e+02 1.4e+02 1.6e+02 57 88 95 4.3 34 60 91 1.2e+02 84 1.2e+02 1.1e+02 78 62 70 1.8e+02 1e+02 72 26 55 1.4e+02 79 1.2e+02 79 98 -22 76 44 1e+02 75 60
- p0(v30): 9.4 1.4e+02 1.3e+02 18 60 1.2e+02 62 53 14 30 63 1.1e+02 1e+02 1.9e+02 1.4e+02 1.7e+02 2e+02 20 1.4e+02 1.8e+02 89 1.1e+02 87 1e+02 75 1.2e+02 1e+02 89 87 1e+02 81 91 1.1e+02 86 1.1e+02 73 92 95 1.2e+02 83 69
- p20(v29): 19 1.5e+02 91 1.2e+02 24 1e+02 1.7e+02 40 62 1e+02 53 1.4e+02 1.3e+02 91 1.7e+02 1.4e+02 1.3e+02 1.4e+02 59 94 1.1e+02 94 86 72 91 96 1e+02 1.2e+02 1e+02 92 1e+02 89 1e+02 1.4e+02 75 1.3e+02 78 1.3e+02 1.2e+02 1.1e+02 75
- p1(v28): 3 1.5e+02 1.1e+02 30 44 1.4e+02 91 39 -6.7 40 68 1.1e+02 1.1e+02 1.9e+02 1.4e+02 1.8e+02 2.1e+02 34 1.3e+02 1.9e+02 1e+02 1.2e+02 67 48 40 1.6e+02 1.1e+02 77 40 69 1.1e+02 57 1.2e+02 1e+02 90 18 70 84 1.3e+02 84 67
- p35(v27): 1.3e+02 75 1.1e+02 48 1.4e+02 1.1e+02 31 1.2e+02 86 68 1.4e+02 69 80 1.5e+02 60 99 1.3e+02 34 1.6e+02 1.5e+02 98 1.2e+02 95 1e+02 84 1.3e+02 95 77 81 94 90 93 1.1e+02 70 1.2e+02 44 1e+02 70 1e+02 77 23
- p25(v26): 49 1e+02 1e+02 46 68 1.2e+02 1.4e+02 30 65 55 1.3e+02 94 86 1e+02 1.6e+02 1.4e+02 1.3e+02 1e+02 1.2e+02 1.4e+02 1e+02 1.1e+02 1.1e+02 94 1.1e+02 63 86 1.1e+02 1.6e+02 1.2e+02 53 86 64 1.3e+02 1.2e+02 1.7e+02 1.1e+02 1.4e+02 1.2e+02 1.1e+02 93
- p98(v25): 1.9e+02 61 61 1.7e+02 1.4e+02 93 1.5e+02 1.3e+02 1.7e+02 1.6e+02 1.6e+02 76 92 19 64 33 16 1.8e+02 70 38 1.3e+02 97 1e+02 73 1.1e+02 1.1e+02 91 1.1e+02 1e+02 81 1.1e+02 91 83 1.3e+02 93 1e+02 92 1e+02 1.1e+02 1.1e+02 1e+02
- p52(v24): 2e+02 47 45 1.6e+02 1.4e+02 1.1e+02 1.9e+02 1e+02 1.7e+02 1.6e+02 2e+02 60 79 -5.3 82 32 6.8 2e+02 79 48 1.3e+02 1.1e+02 1.1e+02 51 1.1e+02 97 82 1.1e+02 1.2e+02 76 99 70 57 1.5e+02 98 1.3e+02 99 1.2e+02 1.1e+02 1.2e+02 1.5e+02
- p84(v23): 1.8e+02 1.1e+02 62 2.3e+02 1.2e+02 77 1.2e+02 1.8e+02 1.6e+02 2.1e+02 72 1.2e+02 1.3e+02 48 33 31 29 1.7e+02 21 2.4 1.3e+02 83 59 51 75 1.8e+02 1.2e+02 1e+02 0.5 44 1.9e+02 95 1.5e+02 1.1e+02 49 -15 45 59 98 84 1.2e+02
- p43(v22): 1.7e+02 1e+02 1.1e+02 2.1e+02 1.3e+02 48 56 2e+02 1.8e+02 1.8e+02 32 1.3e+02 1.3e+02 67 30 36 26 1.3e+02 30 -4.4 1e+02 62 89 1.5e+02 1.4e+02 1e+02 1.1e+02 1.3e+02 76 1.1e+02 1.4e+02 1.6e+02 1.4e+02 73 80 71 79 75 77 77 1.3e+02
- p18(v21): 1.5e+02 1.1e+02 1e+02 2.1e+02 1.2e+02 51 85 1.8e+02 1.8e+02 1.8e+02 42 1.3e+02 1.3e+02 51 53 42 22 1.5e+02 27 -4.2 1e+02 62 97 1.5e+02 1.4e+02 83 1.1e+02 1.3e+02 1e+02 1.2e+02 1.2e+02 1.6e+02 1.3e+02 88 83 1.1e+02 86 93 82 87 1.3e+02
- p16(v20): 1.3e+02 76 46 1.2e+02 89 1.3e+02 2.2e+02 46 1.1e+02 1.2e+02 1.9e+02 70 86 29 1.3e+02 83 65 1.9e+02 92 94 1.3e+02 1.2e+02 97 16 76 1.2e+02 84 1e+02 1.1e+02 62 99 40 57 1.6e+02 90 1.2e+02 89 1.3e+02 1.3e+02 1.2e+02 1e+02
- p5(v19): 1.3e+02 1.2e+02 87 2e+02 93 70 1.1e+02 1.6e+02 1.4e+02 1.8e+02 38 1.3e+02 1.4e+02 68 67 63 54 1.5e+02 26 17 1.2e+02 75 74 95 1e+02 1.4e+02 1.2e+02 1.2e+02 47 79 1.5e+02 1.2e+02 1.4e+02 1e+02 62 47 62 81 95 86 72
- p44(v18): 85 1.3e+02 94 1.6e+02 71 82 1.3e+02 1e+02 1.2e+02 1.4e+02 56 1.3e+02 1.2e+02 72 1.1e+02 95 77 1.4e+02 48 52 1.1e+02 81 92 1.1e+02 1.1e+02 92 1.1e+02 1.3e+02 1e+02 1e+02 1.1e+02 1.2e+02 1.1e+02 1.2e+02 80 1.2e+02 83 1.1e+02 1e+02 1e+02 57
- p51(v17): 1.5e+02 60 89 74 1.4e+02 1.1e+02 96 1e+02 1.1e+02 90 1.7e+02 61 73 92 79 80 87 96 1.4e+02 1.2e+02 1.1e+02 1.2e+02 1.1e+02 87 97 1e+02 86 92 1.2e+02 95 79 84 76 1e+02 1.2e+02 1e+02 1.1e+02 97 1.1e+02 98 38
- p47(v16): 30 1.6e+02 1.1e+02 1.4e+02 41 87 1e+02 99 65 1.1e+02 -1.9 1.5e+02 1.4e+02 1.3e+02 1.2e+02 1.3e+02 1.4e+02 94 52 81 1e+02 83 69 96 85 1.3e+02 1.2e+02 1.1e+02 50 88 1.3e+02 1.1e+02 1.5e+02 98 66 52 62 90 1.1e+02 84 43
- p92(v15): 1.9e+02 56 78 1.6e+02 1.5e+02 78 1.3e+02 1.4e+02 1.9e+02 1.6e+02 1.5e+02 82 86 13 66 30 2.5 1.7e+02 70 27 1.1e+02 86 1.2e+02 1.2e+02 1.4e+02 58 86 1.3e+02 1.5e+02 1.2e+02 85 1.3e+02 73 1.2e+02 1.1e+02 1.7e+02 1.1e+02 1.2e+02 95 1.1e+02 1.6e+02
- p54(v14): 1.6e+02 98 99 1.9e+02 1.3e+02 64 64 1.9e+02 1.6e+02 1.7e+02 59 1.2e+02 1.2e+02 76 35 44 42 1.2e+02 50 22 1.1e+02 75 86 1.3e+02 1.2e+02 1.2e+02 1.1e+02 1.2e+02 67 96 1.4e+02 1.4e+02 1.4e+02 79 83 56 79 72 85 79 79
- p55(v13): 1.5e+02 49 1.2e+02 24 1.6e+02 1.1e+02 37 1.1e+02 1.1e+02 52 1.7e+02 51 58 1.3e+02 69 91 1.1e+02 41 1.8e+02 1.6e+02 92 1.2e+02 1.2e+02 1.3e+02 1.1e+02 82 81 86 1.4e+02 1.2e+02 50 1.1e+02 73 75 1.5e+02 1.2e+02 1.3e+02 93 1e+02 88 85
- p73(v12): 2.1e+02 46 66 1.7e+02 1.6e+02 85 1.3e+02 1.5e+02 1.9e+02 1.7e+02 1.7e+02 70 84 10 49 18 -2.6 1.8e+02 74 29 1.2e+02 93 1.1e+02 97 1.3e+02 88 87 1.2e+02 1.2e+02 95 1e+02 1.1e+02 77 1.2e+02 1e+02 1.3e+02 1e+02 1e+02 99 1.1e+02 1.5e+02
- p48(v11): 1.3e+02 58 1.1e+02 2.8 1.6e+02 1.3e+02 25 1e+02 67 38 1.8e+02 45 62 1.7e+02 63 1.1e+02 1.5e+02 11 1.9e+02 1.9e+02 97 1.4e+02 1e+02 88 72 1.4e+02 87 62 88 91 74 73 90 70 1.4e+02 41 1.1e+02 69 1.1e+02 79 63
- p100(v10): 1.1e+02 99 97 1.1e+02 1e+02 1e+02 1e+02 1e+02 1e+02 1.1e+02 1e+02 98 1e+02 97 96 96 95 1.1e+02 97 96 1.1e+02 1e+02 96 97 1e+02 1.1e+02 98 1.1e+02 1e+02 97 98 1e+02 1e+02 1e+02 1e+02 96 93 99 1.1e+02 95 2.4e+02
- p60(v9): 2e+02 59 84 1.1e+02 1.7e+02 1.1e+02 45 1.7e+02 1.2e+02 1.2e+02 1.5e+02 58 87 1.2e+02 13 50 82 71 1.3e+02 1.1e+02 1.2e+02 1.2e+02 77 63 64 1.8e+02 1e+02 68 25 57 1.4e+02 76 1.2e+02 75 1e+02 -26 77 43 1e+02 73 41
- p74(v8): 0.33 1.6e+02 1.2e+02 79 35 1e+02 91 64 30 70 17 1.4e+02 1.3e+02 1.6e+02 1.4e+02 1.6e+02 1.7e+02 61 90 1.3e+02 95 97 76 97 80 1.2e+02 1.1e+02 1.1e+02 70 97 1.1e+02 1e+02 1.3e+02 97 84 70 74 98 1.1e+02 86 51
- p96(v7): 5.7 1.5e+02 1.4e+02 63 49 98 63 75 39 58 17 1.4e+02 1.2e+02 1.7e+02 1.4e+02 1.6e+02 1.7e+02 42 1e+02 1.4e+02 85 94 87 1.3e+02 98 98 1.1e+02 1.1e+02 95 1.2e+02 89 1.2e+02 1.3e+02 84 99 95 89 1e+02 1.1e+02 82 64
- p81(v6): 2.2e+02 53 46 1.8e+02 1.6e+02 1e+02 1.4e+02 1.5e+02 1.7e+02 1.8e+02 1.8e+02 62 93 29 30 17 17 1.7e+02 76 42 1.4e+02 1.1e+02 81 33 75 1.7e+02 96 90 42 45 1.5e+02 65 1e+02 1.2e+02 81 20 71 70 1.1e+02 97 1.1e+02
- p86(v5): 30 1.5e+02 1.1e+02 1.5e+02 34 69 1.6e+02 70 1.1e+02 1.2e+02 19 1.6e+02 1.3e+02 62 1.6e+02 1.2e+02 82 1.6e+02 32 44 94 65 1.1e+02 1.5e+02 1.5e+02 21 1e+02 1.6e+02 1.7e+02 1.4e+02 74 1.5e+02 94 1.3e+02 89 2.2e+02 1e+02 1.5e+02 1e+02 1.1e+02 1.8e+02
- p19(v4): 46 1.2e+02 57 87 37 1.5e+02 2.2e+02 6.4 36 89 1.4e+02 94 1.1e+02 87 1.7e+02 1.4e+02 1.4e+02 1.5e+02 98 1.4e+02 1.3e+02 1.3e+02 75 -11 40 1.5e+02 95 93 64 45 1.1e+02 19 80 1.6e+02 75 71 68 1.2e+02 1.4e+02 1.2e+02 83
- p94(v3): 1.5e+02 45 87 25 1.5e+02 1.4e+02 84 84 85 58 2.1e+02 35 56 1.2e+02 76 93 1.2e+02 64 1.8e+02 1.7e+02 1.1e+02 1.4e+02 1e+02 60 72 1.3e+02 80 69 1e+02 80 73 54 68 1e+02 1.3e+02 69 1.1e+02 86 1.2e+02 95 59
- p46(v2): 2.4e+02 48 72 1.8e+02 1.9e+02 85 65 2e+02 1.9e+02 1.8e+02 1.5e+02 65 93 58 -9.6 5 17 1.3e+02 85 37 1.2e+02 99 84 81 92 1.6e+02 1e+02 88 39 64 1.5e+02 1e+02 1.2e+02 81 92 2.2 78 50 91 78 1.1e+02
- p79(v1): -34 1.7e+02 1.2e+02 51 9 1.2e+02 1.3e+02 17 1.5 47 32 1.4e+02 1.2e+02 1.6e+02 1.8e+02 1.9e+02 2e+02 68 1e+02 1.6e+02 97 1.1e+02 79 76 71 1.1e+02 1.1e+02 1.1e+02 86 95 93 83 1.1e+02 1.2e+02 86 95 78 1.2e+02 1.3e+02 97 99
The center point is coplanar with a facet, or a vertex is coplanar
with a neighboring facet. The maximum round off error for
computing distances is 1.4e-11. The center point, facets and distances
to the center point are as follows:
center point 115.5 93.65 91.29 106.3 107.1 104.5 107.2 103.4 104.2 107.5 116.2 91.82 98.3 95.25 90.88 91.65 94.38 107.4 101 99.07 109.6 104.7 92.77 84.38 92.46 117 97.12 100.6 90.38 88.37 102.9 91.85 99.54 106 99.45 82.5 90.1 94.75 109.3 94.69 86.16
facet p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.8e-14
facet p14 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.4e-14
facet p14 p13 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -3.6e-14
facet p14 p13 p12 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.8e-14
facet p14 p13 p12 p11 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.1e-14
facet p14 p13 p12 p11 p10 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.6e-14
facet p14 p13 p12 p11 p10 p9 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -6.6e-15
facet p14 p13 p12 p11 p10 p9 p8 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.2e-16
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -5.3e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.7e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -3.9e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -6e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -8.3e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -9.1e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.3e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -9.3e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.1e-13
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -7.9e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -7.4e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.9e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.5e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4.2e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -5e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.5e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p74 p96 p81 p86 p19 p94 p46 p79 distance= -5.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p96 p81 p86 p19 p94 p46 p79 distance= -4.4e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p81 p86 p19 p94 p46 p79 distance= -1.2e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p86 p19 p94 p46 p79 distance= -7.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p19 p94 p46 p79 distance= -3.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p94 p46 p79 distance= -2.7e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p46 p79 distance= -1e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p79 distance= -3.2e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 distance= -1.4e-14
These points either have a maximum or minimum x-coordinate, or
they maximize the determinant for k coordinates. Trial points
are first selected from points that maximize a coordinate.
Because of the high dimension, the min x-coordinate and max-coordinate
points are used if the determinant is non-zero. Option 'Qs' will
do a better, though much slower, job. Instead of 'Qs', you can change
the points by randomly rotating the input with 'QR0'.
The min and max coordinates for each dimension are:
0: -34.17 242.8 difference= 276.9
1: 41.16 170.2 difference= 129
2: 44.87 144.1 difference= 99.26
3: -8.179 233 difference= 241.1
4: 8.965 188.5 difference= 179.5
5: 47.84 145.2 difference= 97.33
6: -2.737 218.1 difference= 220.9
7: -12.18 204.8 difference= 217
8: -6.742 198.8 difference= 205.6
9: 7.474 206.9 difference= 199.4
10: -19.73 213.7 difference= 233.4
11: 34.9 158.6 difference= 123.7
12: 56.27 147.8 difference= 91.56
13: -5.28 206.4 difference= 211.7
14: -9.578 192.3 difference= 201.9
15: 4.992 190.1 difference= 185.2
16: -4.608 209.4 difference= 214
17: 2.683 204.3 difference= 201.6
18: 21 194.6 difference= 173.6
19: -4.403 199.5 difference= 203.9
20: 80.48 136.5 difference= 56.07
21: 62.07 138.9 difference= 76.82
22: 59.19 129.1 difference= 69.96
23: -10.94 185.3 difference= 196.2
24: 40.24 152.6 difference= 112.3
25: 21.15 183.6 difference= 162.5
26: 78.41 120.4 difference= 41.96
27: 62.19 156.4 difference= 94.22
28: 0.4971 179.8 difference= 179.3
29: 43.8 143.4 difference= 99.65
30: 36.92 185.1 difference= 148.2
31: 19.33 178.3 difference= 159
32: 44.23 156.2 difference= 112
33: 51.54 163.7 difference= 112.1
34: 48.53 150 difference= 101.5
35: -26 220.3 difference= 246.3
36: 44.64 133.6 difference= 88.94
37: 43.41 154.3 difference= 110.8
38: 75.61 142.4 difference= 66.82
39: 64.55 127.9 difference= 63.31
40: 0 242.8 difference= 242.8
If the input should be full dimensional, you have several options that
may determine an initial simplex:
- use 'QJ' to joggle the input and make it full dimensional
- use 'QbB' to scale the points to the unit cube
- use 'QR0' to randomly rotate the input for different maximum points
- use 'Qs' to search all points for the initial simplex
- use 'En' to specify a maximum roundoff error less than 1.4e-11.
- trace execution with 'T3' to see the determinant for each point.
If the input is lower dimensional:
- use 'QJ' to joggle the input and make it full dimensional
- use 'Qbk:0Bk:0' to delete coordinate k from the input. You should
pick the coordinate with the least range. The hull will have the
correct topology.
- determine the flat containing the points, rotate the points
into a coordinate plane, and delete the other coordinates.
- add one or more points to make the input full dimensional.
"""
The above exception was the direct cause of the following exception:
QhullError Traceback (most recent call last)
Cell In[100], line 12
7 from gtda.homology import WeakAlphaPersistence
10 wa_pers = WeakAlphaPersistence(homology_dimensions=(0, 1, 2), n_jobs=-1)
---> 12 diagrams_wa_pers = wa_pers.fit_transform([X_pca, X_pls])
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/gtda/utils/_docs.py:106, in adapt_fit_transform_docs.<locals>.make_new_fit_transform.<locals>.fit_transform_wrapper(*args, **kwargs)
104 @wraps(original_fit_transform)
105 def fit_transform_wrapper(*args, **kwargs):
--> 106 return original_fit_transform(*args, **kwargs)
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/sklearn/utils/_set_output.py:295, in _wrap_method_output.<locals>.wrapped(self, X, *args, **kwargs)
293 @wraps(f)
294 def wrapped(self, X, *args, **kwargs):
--> 295 data_to_wrap = f(self, X, *args, **kwargs)
296 if isinstance(data_to_wrap, tuple):
297 # only wrap the first output for cross decomposition
298 return_tuple = (
299 _wrap_data_with_container(method, data_to_wrap[0], X, self),
300 *data_to_wrap[1:],
301 )
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/sklearn/base.py:1098, in TransformerMixin.fit_transform(self, X, y, **fit_params)
1083 warnings.warn(
1084 (
1085 f"This object ({self.__class__.__name__}) has a `transform`"
(...)
1093 UserWarning,
1094 )
1096 if y is None:
1097 # fit method of arity 1 (unsupervised transformation)
-> 1098 return self.fit(X, **fit_params).transform(X)
1099 else:
1100 # fit method of arity 2 (supervised transformation)
1101 return self.fit(X, y, **fit_params).transform(X)
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/sklearn/utils/_set_output.py:295, in _wrap_method_output.<locals>.wrapped(self, X, *args, **kwargs)
293 @wraps(f)
294 def wrapped(self, X, *args, **kwargs):
--> 295 data_to_wrap = f(self, X, *args, **kwargs)
296 if isinstance(data_to_wrap, tuple):
297 # only wrap the first output for cross decomposition
298 return_tuple = (
299 _wrap_data_with_container(method, data_to_wrap[0], X, self),
300 *data_to_wrap[1:],
301 )
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/gtda/homology/simplicial.py:1208, in WeakAlphaPersistence.transform(self, X, y)
1205 check_is_fitted(self)
1206 X = check_point_clouds(X)
-> 1208 Xt = Parallel(n_jobs=self.n_jobs)(
1209 delayed(self._weak_alpha_diagram)(x) for x in X)
1211 Xt = _postprocess_diagrams(
1212 Xt, "ripser", self._homology_dimensions, self.infinity_values_,
1213 self.reduced_homology
1214 )
1215 return Xt
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py:1952, in Parallel.__call__(self, iterable)
1946 # The first item from the output is blank, but it makes the interpreter
1947 # progress until it enters the Try/Except block of the generator and
1948 # reach the first `yield` statement. This starts the aynchronous
1949 # dispatch of the tasks to the workers.
1950 next(output)
-> 1952 return output if self.return_generator else list(output)
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py:1595, in Parallel._get_outputs(self, iterator, pre_dispatch)
1592 yield
1594 with self._backend.retrieval_context():
-> 1595 yield from self._retrieve()
1597 except GeneratorExit:
1598 # The generator has been garbage collected before being fully
1599 # consumed. This aborts the remaining tasks if possible and warn
1600 # the user if necessary.
1601 self._exception = True
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py:1699, in Parallel._retrieve(self)
1692 while self._wait_retrieval():
1693
1694 # If the callback thread of a worker has signaled that its task
1695 # triggered an exception, or if the retrieval loop has raised an
1696 # exception (e.g. `GeneratorExit`), exit the loop and surface the
1697 # worker traceback.
1698 if self._aborting:
-> 1699 self._raise_error_fast()
1700 break
1702 # If the next job is not ready for retrieval yet, we just wait for
1703 # async callbacks to progress.
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py:1734, in Parallel._raise_error_fast(self)
1730 # If this error job exists, immediatly raise the error by
1731 # calling get_result. This job might not exists if abort has been
1732 # called directly or if the generator is gc'ed.
1733 if error_job is not None:
-> 1734 error_job.get_result(self.timeout)
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py:736, in BatchCompletionCallBack.get_result(self, timeout)
730 backend = self.parallel._backend
732 if backend.supports_retrieve_callback:
733 # We assume that the result has already been retrieved by the
734 # callback thread, and is stored internally. It's just waiting to
735 # be returned.
--> 736 return self._return_or_raise()
738 # For other backends, the main thread needs to run the retrieval step.
739 try:
File ~/miniconda3/envs/neurometry/lib/python3.9/site-packages/joblib/parallel.py:754, in BatchCompletionCallBack._return_or_raise(self)
752 try:
753 if self.status == TASK_ERROR:
--> 754 raise self._result
755 return self._result
756 finally:
QhullError: QH6154 Qhull precision error: Initial simplex is flat (facet 1 is coplanar with the interior point)
While executing: | qhull d Qt Q12 Qx Qc Qz Qbb
Options selected for Qhull 2019.1.r 2019/06/21:
run-id 1336544124 delaunay Qtriangulate Q12-allow-wide Qxact-merge
Qcoplanar-keep Qz-infinity-point Qbbound-last _zero-centrum
Qinterior-keep Q3-no-merge-vertices-dim-high Pgood _max-width 2.8e+02
Error-roundoff 1.4e-11 _one-merge 1.2e-09 Visible-distance 8.6e-11
U-max-coplanar 8.6e-11 Width-outside 1.7e-10 _wide-facet 5.2e-10
_maxoutside 1.2e-09
precision problems (corrected unless 'Q0' or an error)
42 nearly singular or axis-parallel hyperplanes
7 zero divisors during gaussian elimination
The input to qhull appears to be less than 41 dimensional, or a
computation has overflowed.
Qhull could not construct a clearly convex simplex from points:
- p14(v42): 1.6e+02 68 1e+02 91 1.6e+02 1e+02 40 1.5e+02 1.2e+02 1e+02 1.3e+02 71 85 1.2e+02 40 69 90 64 1.3e+02 1.1e+02 1e+02 1.1e+02 95 1.1e+02 95 1.3e+02 96 85 78 92 1e+02 1e+02 1.1e+02 73 1.2e+02 45 97 68 98 78 23
- p13(v41): 2e+02 41 64 1.1e+02 1.6e+02 1.1e+02 1.3e+02 1.2e+02 1.5e+02 1.3e+02 2e+02 48 71 51 56 44 45 1.4e+02 1.2e+02 91 1.2e+02 1.2e+02 1e+02 60 91 1.2e+02 84 90 99 74 99 69 72 1.2e+02 1.1e+02 82 1e+02 91 1.1e+02 1e+02 71
- p12(v40): -3.9 1.4e+02 1.1e+02 22 40 1.3e+02 1.1e+02 24 1.3 32 77 1.1e+02 1.1e+02 1.7e+02 1.6e+02 1.8e+02 2e+02 47 1.4e+02 1.8e+02 98 1.2e+02 82 67 61 1.3e+02 1e+02 89 81 88 86 68 1e+02 1.1e+02 1e+02 74 85 1e+02 1.3e+02 93 76
- p11(v39): 1.9e+02 53 66 1.4e+02 1.5e+02 1e+02 1.3e+02 1.3e+02 1.6e+02 1.4e+02 1.8e+02 65 82 37 61 41 32 1.5e+02 95 64 1.2e+02 1.1e+02 1e+02 75 1e+02 1.1e+02 88 1e+02 1.1e+02 83 1e+02 86 78 1.2e+02 1e+02 1e+02 98 98 1.1e+02 1e+02 80
- p10(v38): 1.9e+02 54 1.2e+02 1.1e+02 1.8e+02 70 3.5 1.9e+02 1.7e+02 1.2e+02 1.2e+02 78 81 1e+02 19 42 50 65 1.2e+02 70 89 87 1.2e+02 1.8e+02 1.5e+02 74 93 1.1e+02 1.3e+02 1.3e+02 80 1.6e+02 1.1e+02 52 1.3e+02 1.1e+02 1.2e+02 77 79 76 1.2e+02
- p9(v37): 64 98 82 68 65 1.2e+02 1.8e+02 22 77 73 1.5e+02 89 86 72 1.6e+02 1.3e+02 1.1e+02 1.4e+02 1.1e+02 1.3e+02 1.1e+02 1.1e+02 1.1e+02 71 1e+02 66 84 1.1e+02 1.6e+02 1e+02 59 72 54 1.5e+02 1.1e+02 1.8e+02 1.1e+02 1.4e+02 1.3e+02 1.2e+02 1.1e+02
- p8(v36): 1e+02 79 97 54 1.1e+02 1.2e+02 1.2e+02 67 90 68 1.5e+02 74 77 99 1.2e+02 1.1e+02 1.1e+02 96 1.4e+02 1.4e+02 1e+02 1.2e+02 1.1e+02 95 1e+02 81 85 1e+02 1.4e+02 1.1e+02 62 86 68 1.1e+02 1.2e+02 1.4e+02 1.1e+02 1.2e+02 1.2e+02 1e+02 55
- p7(v35): 32 1.3e+02 1.2e+02 33 68 1.3e+02 73 60 18 45 81 1e+02 1e+02 1.8e+02 1.2e+02 1.6e+02 1.9e+02 32 1.4e+02 1.8e+02 99 1.2e+02 76 69 56 1.5e+02 1e+02 80 57 80 1e+02 71 1.2e+02 92 1e+02 34 80 83 1.2e+02 82 39
- p6(v34): 26 1.3e+02 86 44 46 1.4e+02 1.5e+02 16 13 54 1.2e+02 95 1e+02 1.4e+02 1.6e+02 1.6e+02 1.8e+02 86 1.3e+02 1.7e+02 1.1e+02 1.3e+02 76 20 42 1.5e+02 98 83 63 61 1e+02 36 93 1.3e+02 90 56 76 1e+02 1.4e+02 1e+02 59
- p4(v33): 1.2e+02 69 83 45 1.2e+02 1.4e+02 1.1e+02 69 74 68 1.8e+02 55 72 1.1e+02 98 1.1e+02 1.2e+02 83 1.5e+02 1.6e+02 1.1e+02 1.3e+02 96 49 67 1.3e+02 86 77 92 75 85 52 75 1.1e+02 1.2e+02 69 99 93 1.2e+02 99 32
- p3(v32): 1.4e+02 76 68 1.1e+02 1.1e+02 1.2e+02 1.5e+02 86 1.2e+02 1.2e+02 1.6e+02 75 88 59 98 77 71 1.5e+02 1e+02 94 1.2e+02 1.1e+02 97 54 87 1.2e+02 89 1e+02 1e+02 76 1e+02 68 77 1.3e+02 98 98 92 1.1e+02 1.2e+02 1.1e+02 42
- p2(v31): 2.1e+02 53 77 1.3e+02 1.8e+02 1e+02 56 1.8e+02 1.4e+02 1.4e+02 1.6e+02 57 88 95 4.3 34 60 91 1.2e+02 84 1.2e+02 1.1e+02 78 62 70 1.8e+02 1e+02 72 26 55 1.4e+02 79 1.2e+02 79 98 -22 76 44 1e+02 75 60
- p0(v30): 9.4 1.4e+02 1.3e+02 18 60 1.2e+02 62 53 14 30 63 1.1e+02 1e+02 1.9e+02 1.4e+02 1.7e+02 2e+02 20 1.4e+02 1.8e+02 89 1.1e+02 87 1e+02 75 1.2e+02 1e+02 89 87 1e+02 81 91 1.1e+02 86 1.1e+02 73 92 95 1.2e+02 83 69
- p20(v29): 19 1.5e+02 91 1.2e+02 24 1e+02 1.7e+02 40 62 1e+02 53 1.4e+02 1.3e+02 91 1.7e+02 1.4e+02 1.3e+02 1.4e+02 59 94 1.1e+02 94 86 72 91 96 1e+02 1.2e+02 1e+02 92 1e+02 89 1e+02 1.4e+02 75 1.3e+02 78 1.3e+02 1.2e+02 1.1e+02 75
- p1(v28): 3 1.5e+02 1.1e+02 30 44 1.4e+02 91 39 -6.7 40 68 1.1e+02 1.1e+02 1.9e+02 1.4e+02 1.8e+02 2.1e+02 34 1.3e+02 1.9e+02 1e+02 1.2e+02 67 48 40 1.6e+02 1.1e+02 77 40 69 1.1e+02 57 1.2e+02 1e+02 90 18 70 84 1.3e+02 84 67
- p35(v27): 1.3e+02 75 1.1e+02 48 1.4e+02 1.1e+02 31 1.2e+02 86 68 1.4e+02 69 80 1.5e+02 60 99 1.3e+02 34 1.6e+02 1.5e+02 98 1.2e+02 95 1e+02 84 1.3e+02 95 77 81 94 90 93 1.1e+02 70 1.2e+02 44 1e+02 70 1e+02 77 23
- p25(v26): 49 1e+02 1e+02 46 68 1.2e+02 1.4e+02 30 65 55 1.3e+02 94 86 1e+02 1.6e+02 1.4e+02 1.3e+02 1e+02 1.2e+02 1.4e+02 1e+02 1.1e+02 1.1e+02 94 1.1e+02 63 86 1.1e+02 1.6e+02 1.2e+02 53 86 64 1.3e+02 1.2e+02 1.7e+02 1.1e+02 1.4e+02 1.2e+02 1.1e+02 93
- p98(v25): 1.9e+02 61 61 1.7e+02 1.4e+02 93 1.5e+02 1.3e+02 1.7e+02 1.6e+02 1.6e+02 76 92 19 64 33 16 1.8e+02 70 38 1.3e+02 97 1e+02 73 1.1e+02 1.1e+02 91 1.1e+02 1e+02 81 1.1e+02 91 83 1.3e+02 93 1e+02 92 1e+02 1.1e+02 1.1e+02 1e+02
- p52(v24): 2e+02 47 45 1.6e+02 1.4e+02 1.1e+02 1.9e+02 1e+02 1.7e+02 1.6e+02 2e+02 60 79 -5.3 82 32 6.8 2e+02 79 48 1.3e+02 1.1e+02 1.1e+02 51 1.1e+02 97 82 1.1e+02 1.2e+02 76 99 70 57 1.5e+02 98 1.3e+02 99 1.2e+02 1.1e+02 1.2e+02 1.5e+02
- p84(v23): 1.8e+02 1.1e+02 62 2.3e+02 1.2e+02 77 1.2e+02 1.8e+02 1.6e+02 2.1e+02 72 1.2e+02 1.3e+02 48 33 31 29 1.7e+02 21 2.4 1.3e+02 83 59 51 75 1.8e+02 1.2e+02 1e+02 0.5 44 1.9e+02 95 1.5e+02 1.1e+02 49 -15 45 59 98 84 1.2e+02
- p43(v22): 1.7e+02 1e+02 1.1e+02 2.1e+02 1.3e+02 48 56 2e+02 1.8e+02 1.8e+02 32 1.3e+02 1.3e+02 67 30 36 26 1.3e+02 30 -4.4 1e+02 62 89 1.5e+02 1.4e+02 1e+02 1.1e+02 1.3e+02 76 1.1e+02 1.4e+02 1.6e+02 1.4e+02 73 80 71 79 75 77 77 1.3e+02
- p18(v21): 1.5e+02 1.1e+02 1e+02 2.1e+02 1.2e+02 51 85 1.8e+02 1.8e+02 1.8e+02 42 1.3e+02 1.3e+02 51 53 42 22 1.5e+02 27 -4.2 1e+02 62 97 1.5e+02 1.4e+02 83 1.1e+02 1.3e+02 1e+02 1.2e+02 1.2e+02 1.6e+02 1.3e+02 88 83 1.1e+02 86 93 82 87 1.3e+02
- p16(v20): 1.3e+02 76 46 1.2e+02 89 1.3e+02 2.2e+02 46 1.1e+02 1.2e+02 1.9e+02 70 86 29 1.3e+02 83 65 1.9e+02 92 94 1.3e+02 1.2e+02 97 16 76 1.2e+02 84 1e+02 1.1e+02 62 99 40 57 1.6e+02 90 1.2e+02 89 1.3e+02 1.3e+02 1.2e+02 1e+02
- p5(v19): 1.3e+02 1.2e+02 87 2e+02 93 70 1.1e+02 1.6e+02 1.4e+02 1.8e+02 38 1.3e+02 1.4e+02 68 67 63 54 1.5e+02 26 17 1.2e+02 75 74 95 1e+02 1.4e+02 1.2e+02 1.2e+02 47 79 1.5e+02 1.2e+02 1.4e+02 1e+02 62 47 62 81 95 86 72
- p44(v18): 85 1.3e+02 94 1.6e+02 71 82 1.3e+02 1e+02 1.2e+02 1.4e+02 56 1.3e+02 1.2e+02 72 1.1e+02 95 77 1.4e+02 48 52 1.1e+02 81 92 1.1e+02 1.1e+02 92 1.1e+02 1.3e+02 1e+02 1e+02 1.1e+02 1.2e+02 1.1e+02 1.2e+02 80 1.2e+02 83 1.1e+02 1e+02 1e+02 57
- p51(v17): 1.5e+02 60 89 74 1.4e+02 1.1e+02 96 1e+02 1.1e+02 90 1.7e+02 61 73 92 79 80 87 96 1.4e+02 1.2e+02 1.1e+02 1.2e+02 1.1e+02 87 97 1e+02 86 92 1.2e+02 95 79 84 76 1e+02 1.2e+02 1e+02 1.1e+02 97 1.1e+02 98 38
- p47(v16): 30 1.6e+02 1.1e+02 1.4e+02 41 87 1e+02 99 65 1.1e+02 -1.9 1.5e+02 1.4e+02 1.3e+02 1.2e+02 1.3e+02 1.4e+02 94 52 81 1e+02 83 69 96 85 1.3e+02 1.2e+02 1.1e+02 50 88 1.3e+02 1.1e+02 1.5e+02 98 66 52 62 90 1.1e+02 84 43
- p92(v15): 1.9e+02 56 78 1.6e+02 1.5e+02 78 1.3e+02 1.4e+02 1.9e+02 1.6e+02 1.5e+02 82 86 13 66 30 2.5 1.7e+02 70 27 1.1e+02 86 1.2e+02 1.2e+02 1.4e+02 58 86 1.3e+02 1.5e+02 1.2e+02 85 1.3e+02 73 1.2e+02 1.1e+02 1.7e+02 1.1e+02 1.2e+02 95 1.1e+02 1.6e+02
- p54(v14): 1.6e+02 98 99 1.9e+02 1.3e+02 64 64 1.9e+02 1.6e+02 1.7e+02 59 1.2e+02 1.2e+02 76 35 44 42 1.2e+02 50 22 1.1e+02 75 86 1.3e+02 1.2e+02 1.2e+02 1.1e+02 1.2e+02 67 96 1.4e+02 1.4e+02 1.4e+02 79 83 56 79 72 85 79 79
- p55(v13): 1.5e+02 49 1.2e+02 24 1.6e+02 1.1e+02 37 1.1e+02 1.1e+02 52 1.7e+02 51 58 1.3e+02 69 91 1.1e+02 41 1.8e+02 1.6e+02 92 1.2e+02 1.2e+02 1.3e+02 1.1e+02 82 81 86 1.4e+02 1.2e+02 50 1.1e+02 73 75 1.5e+02 1.2e+02 1.3e+02 93 1e+02 88 85
- p73(v12): 2.1e+02 46 66 1.7e+02 1.6e+02 85 1.3e+02 1.5e+02 1.9e+02 1.7e+02 1.7e+02 70 84 10 49 18 -2.6 1.8e+02 74 29 1.2e+02 93 1.1e+02 97 1.3e+02 88 87 1.2e+02 1.2e+02 95 1e+02 1.1e+02 77 1.2e+02 1e+02 1.3e+02 1e+02 1e+02 99 1.1e+02 1.5e+02
- p48(v11): 1.3e+02 58 1.1e+02 2.8 1.6e+02 1.3e+02 25 1e+02 67 38 1.8e+02 45 62 1.7e+02 63 1.1e+02 1.5e+02 11 1.9e+02 1.9e+02 97 1.4e+02 1e+02 88 72 1.4e+02 87 62 88 91 74 73 90 70 1.4e+02 41 1.1e+02 69 1.1e+02 79 63
- p100(v10): 1.1e+02 99 97 1.1e+02 1e+02 1e+02 1e+02 1e+02 1e+02 1.1e+02 1e+02 98 1e+02 97 96 96 95 1.1e+02 97 96 1.1e+02 1e+02 96 97 1e+02 1.1e+02 98 1.1e+02 1e+02 97 98 1e+02 1e+02 1e+02 1e+02 96 93 99 1.1e+02 95 2.4e+02
- p60(v9): 2e+02 59 84 1.1e+02 1.7e+02 1.1e+02 45 1.7e+02 1.2e+02 1.2e+02 1.5e+02 58 87 1.2e+02 13 50 82 71 1.3e+02 1.1e+02 1.2e+02 1.2e+02 77 63 64 1.8e+02 1e+02 68 25 57 1.4e+02 76 1.2e+02 75 1e+02 -26 77 43 1e+02 73 41
- p74(v8): 0.33 1.6e+02 1.2e+02 79 35 1e+02 91 64 30 70 17 1.4e+02 1.3e+02 1.6e+02 1.4e+02 1.6e+02 1.7e+02 61 90 1.3e+02 95 97 76 97 80 1.2e+02 1.1e+02 1.1e+02 70 97 1.1e+02 1e+02 1.3e+02 97 84 70 74 98 1.1e+02 86 51
- p96(v7): 5.7 1.5e+02 1.4e+02 63 49 98 63 75 39 58 17 1.4e+02 1.2e+02 1.7e+02 1.4e+02 1.6e+02 1.7e+02 42 1e+02 1.4e+02 85 94 87 1.3e+02 98 98 1.1e+02 1.1e+02 95 1.2e+02 89 1.2e+02 1.3e+02 84 99 95 89 1e+02 1.1e+02 82 64
- p81(v6): 2.2e+02 53 46 1.8e+02 1.6e+02 1e+02 1.4e+02 1.5e+02 1.7e+02 1.8e+02 1.8e+02 62 93 29 30 17 17 1.7e+02 76 42 1.4e+02 1.1e+02 81 33 75 1.7e+02 96 90 42 45 1.5e+02 65 1e+02 1.2e+02 81 20 71 70 1.1e+02 97 1.1e+02
- p86(v5): 30 1.5e+02 1.1e+02 1.5e+02 34 69 1.6e+02 70 1.1e+02 1.2e+02 19 1.6e+02 1.3e+02 62 1.6e+02 1.2e+02 82 1.6e+02 32 44 94 65 1.1e+02 1.5e+02 1.5e+02 21 1e+02 1.6e+02 1.7e+02 1.4e+02 74 1.5e+02 94 1.3e+02 89 2.2e+02 1e+02 1.5e+02 1e+02 1.1e+02 1.8e+02
- p19(v4): 46 1.2e+02 57 87 37 1.5e+02 2.2e+02 6.4 36 89 1.4e+02 94 1.1e+02 87 1.7e+02 1.4e+02 1.4e+02 1.5e+02 98 1.4e+02 1.3e+02 1.3e+02 75 -11 40 1.5e+02 95 93 64 45 1.1e+02 19 80 1.6e+02 75 71 68 1.2e+02 1.4e+02 1.2e+02 83
- p94(v3): 1.5e+02 45 87 25 1.5e+02 1.4e+02 84 84 85 58 2.1e+02 35 56 1.2e+02 76 93 1.2e+02 64 1.8e+02 1.7e+02 1.1e+02 1.4e+02 1e+02 60 72 1.3e+02 80 69 1e+02 80 73 54 68 1e+02 1.3e+02 69 1.1e+02 86 1.2e+02 95 59
- p46(v2): 2.4e+02 48 72 1.8e+02 1.9e+02 85 65 2e+02 1.9e+02 1.8e+02 1.5e+02 65 93 58 -9.6 5 17 1.3e+02 85 37 1.2e+02 99 84 81 92 1.6e+02 1e+02 88 39 64 1.5e+02 1e+02 1.2e+02 81 92 2.2 78 50 91 78 1.1e+02
- p79(v1): -34 1.7e+02 1.2e+02 51 9 1.2e+02 1.3e+02 17 1.5 47 32 1.4e+02 1.2e+02 1.6e+02 1.8e+02 1.9e+02 2e+02 68 1e+02 1.6e+02 97 1.1e+02 79 76 71 1.1e+02 1.1e+02 1.1e+02 86 95 93 83 1.1e+02 1.2e+02 86 95 78 1.2e+02 1.3e+02 97 99
The center point is coplanar with a facet, or a vertex is coplanar
with a neighboring facet. The maximum round off error for
computing distances is 1.4e-11. The center point, facets and distances
to the center point are as follows:
center point 115.5 93.65 91.29 106.3 107.1 104.5 107.2 103.4 104.2 107.5 116.2 91.82 98.3 95.25 90.88 91.65 94.38 107.4 101 99.07 109.6 104.7 92.77 84.38 92.46 117 97.12 100.6 90.38 88.37 102.9 91.85 99.54 106 99.45 82.5 90.1 94.75 109.3 94.69 86.16
facet p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.8e-14
facet p14 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.4e-14
facet p14 p13 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -3.6e-14
facet p14 p13 p12 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.8e-14
facet p14 p13 p12 p11 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.1e-14
facet p14 p13 p12 p11 p10 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.6e-14
facet p14 p13 p12 p11 p10 p9 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -6.6e-15
facet p14 p13 p12 p11 p10 p9 p8 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.2e-16
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -5.3e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.7e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -3.9e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -6e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -8.3e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -9.1e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.6e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.3e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -9.3e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.1e-13
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -7.9e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -7.4e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.9e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -2.5e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -4.2e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -5e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p100 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.5e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p60 p74 p96 p81 p86 p19 p94 p46 p79 distance= -1.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p74 p96 p81 p86 p19 p94 p46 p79 distance= -5.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p96 p81 p86 p19 p94 p46 p79 distance= -4.4e-15
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p81 p86 p19 p94 p46 p79 distance= -1.2e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p86 p19 p94 p46 p79 distance= -7.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p19 p94 p46 p79 distance= -3.4e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p94 p46 p79 distance= -2.7e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p46 p79 distance= -1e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p79 distance= -3.2e-14
facet p14 p13 p12 p11 p10 p9 p8 p7 p6 p4 p3 p2 p0 p20 p1 p35 p25 p98 p52 p84 p43 p18 p16 p5 p44 p51 p47 p92 p54 p55 p73 p48 p100 p60 p74 p96 p81 p86 p19 p94 p46 distance= -1.4e-14
These points either have a maximum or minimum x-coordinate, or
they maximize the determinant for k coordinates. Trial points
are first selected from points that maximize a coordinate.
Because of the high dimension, the min x-coordinate and max-coordinate
points are used if the determinant is non-zero. Option 'Qs' will
do a better, though much slower, job. Instead of 'Qs', you can change
the points by randomly rotating the input with 'QR0'.
The min and max coordinates for each dimension are:
0: -34.17 242.8 difference= 276.9
1: 41.16 170.2 difference= 129
2: 44.87 144.1 difference= 99.26
3: -8.179 233 difference= 241.1
4: 8.965 188.5 difference= 179.5
5: 47.84 145.2 difference= 97.33
6: -2.737 218.1 difference= 220.9
7: -12.18 204.8 difference= 217
8: -6.742 198.8 difference= 205.6
9: 7.474 206.9 difference= 199.4
10: -19.73 213.7 difference= 233.4
11: 34.9 158.6 difference= 123.7
12: 56.27 147.8 difference= 91.56
13: -5.28 206.4 difference= 211.7
14: -9.578 192.3 difference= 201.9
15: 4.992 190.1 difference= 185.2
16: -4.608 209.4 difference= 214
17: 2.683 204.3 difference= 201.6
18: 21 194.6 difference= 173.6
19: -4.403 199.5 difference= 203.9
20: 80.48 136.5 difference= 56.07
21: 62.07 138.9 difference= 76.82
22: 59.19 129.1 difference= 69.96
23: -10.94 185.3 difference= 196.2
24: 40.24 152.6 difference= 112.3
25: 21.15 183.6 difference= 162.5
26: 78.41 120.4 difference= 41.96
27: 62.19 156.4 difference= 94.22
28: 0.4971 179.8 difference= 179.3
29: 43.8 143.4 difference= 99.65
30: 36.92 185.1 difference= 148.2
31: 19.33 178.3 difference= 159
32: 44.23 156.2 difference= 112
33: 51.54 163.7 difference= 112.1
34: 48.53 150 difference= 101.5
35: -26 220.3 difference= 246.3
36: 44.64 133.6 difference= 88.94
37: 43.41 154.3 difference= 110.8
38: 75.61 142.4 difference= 66.82
39: 64.55 127.9 difference= 63.31
40: 0 242.8 difference= 242.8
If the input should be full dimensional, you have several options that
may determine an initial simplex:
- use 'QJ' to joggle the input and make it full dimensional
- use 'QbB' to scale the points to the unit cube
- use 'QR0' to randomly rotate the input for different maximum points
- use 'Qs' to search all points for the initial simplex
- use 'En' to specify a maximum roundoff error less than 1.4e-11.
- trace execution with 'T3' to see the determinant for each point.
If the input is lower dimensional:
- use 'QJ' to joggle the input and make it full dimensional
- use 'Qbk:0Bk:0' to delete coordinate k from the input. You should
pick the coordinate with the least range. The hull will have the
correct topology.
- determine the flat containing the points, rotate the points
into a coordinate plane, and delete the other coordinates.
- add one or more points to make the input full dimensional.