Notebook source code: notebooks/04_methods_estimate_manifold_geometry.ipynb
Classify Neural Manifold Topology#
Set-up + Imports#
In [2]:
import setup
setup.main()
%load_ext autoreload
%autoreload 2
%load_ext jupyter_black
import os
import matplotlib.pyplot as plt
import numpy as np
import torch
os.environ["GEOMSTATS_BACKEND"] = "pytorch"
import geomstats.backend as gs
import neurometry.datasets.synthetic as synthetic
Working directory: /home/facosta/neurometry/neurometry
Directory added to path: /home/facosta/neurometry
Directory added to path: /home/facosta/neurometry/neurometry
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
The jupyter_black extension is already loaded. To reload it, use:
%reload_ext jupyter_black
In [59]:
import neurometry.datasets.synthetic as synthetic
circle_task_points = synthetic.hypersphere(1, 1000)
circle_noisy_points, circle_manifold_points = synthetic.synthetic_neural_manifold(
points=circle_task_points,
encoding_dim=10,
nonlinearity="sigmoid",
scales=gs.ones(10),
poisson_multiplier=100,
)
noise level: 0.71%
In [60]:
# Plot noisy_points and manifold_points in 3D with plotly
import plotly.graph_objects as go
fig = go.Figure()
fig.add_trace(
go.Scatter3d(
x=circle_noisy_points[:, 0],
y=circle_noisy_points[:, 1],
z=circle_noisy_points[:, 2],
mode="markers",
marker=dict(size=3),
)
)
fig.add_trace(
go.Scatter3d(
x=circle_manifold_points[:, 0],
y=circle_manifold_points[:, 1],
z=circle_manifold_points[:, 2],
mode="markers",
marker=dict(size=3),
)
)
fig.show()
In [61]:
from neurometry.estimators.topology.persistent_homology import compute_diagrams_shuffle
import neurometry.datasets.synthetic as synthetic
circle_task_points = synthetic.hypersphere(1, 1000)
circle_noisy_points, circle_manifold_points = synthetic.synthetic_neural_manifold(
points=circle_task_points,
encoding_dim=10,
nonlinearity="sigmoid",
scales=gs.ones(10),
poisson_multiplier=100,
)
num_shuffles = 100
circle_noisy_diagrams = compute_diagrams_shuffle(
circle_noisy_points, num_shuffles=num_shuffles, homology_dimensions=[0, 1, 2]
)
from gtda.diagrams import PersistenceEntropy
circle_PE = PersistenceEntropy()
circle_features = circle_PE.fit_transform(circle_noisy_diagrams)
# manifold_diagrams = compute_diagrams_shuffle(manifold_points, num_shuffles=num_shuffles)
In [20]:
X = noisy_points
def _shuffle_entries(data, rng):
return np.array([rng.permutation(row) for row in data])
seed = 0
rng = np.random.default_rng(seed)
shuffled_Xs = [_shuffle_entries(X, rng) for _ in range(num_shuffles)]
In [21]:
import plotly.graph_objects as go
fig = go.Figure()
fig.add_trace(
go.Scatter3d(
x=X[:, 0],
y=X[:, 1],
z=X[:, 2],
mode="markers",
marker=dict(size=3),
)
)
i = 5
fig.add_trace(
go.Scatter3d(
x=shuffled_Xs[i][:, 0],
y=shuffled_Xs[i][:, 1],
z=shuffled_Xs[i][:, 2],
mode="markers",
marker=dict(size=3),
)
)
In [33]:
from gtda.plotting import plot_diagram
plot_diagram(noisy_diagrams[0])
In [34]:
plot_diagram(noisy_diagrams[70])
In [62]:
from gtda.diagrams import PersistenceEntropy
circle_PE = PersistenceEntropy()
circle_features = circle_PE.fit_transform(circle_noisy_diagrams)
In [40]:
# 3d plot with plotly
fig = go.Figure()
fig.add_trace(
go.Scatter3d(
x=features[1:, 0],
y=features[1:, 1],
z=features[1:, 2],
mode="markers",
marker=dict(size=3, color="blue"),
)
)
fig.add_trace(
go.Scatter3d(
x=[features[0, 0]],
y=[features[0, 1]],
z=[features[0, 2]],
mode="markers",
marker=dict(size=3, color="red"),
)
)
In [9]:
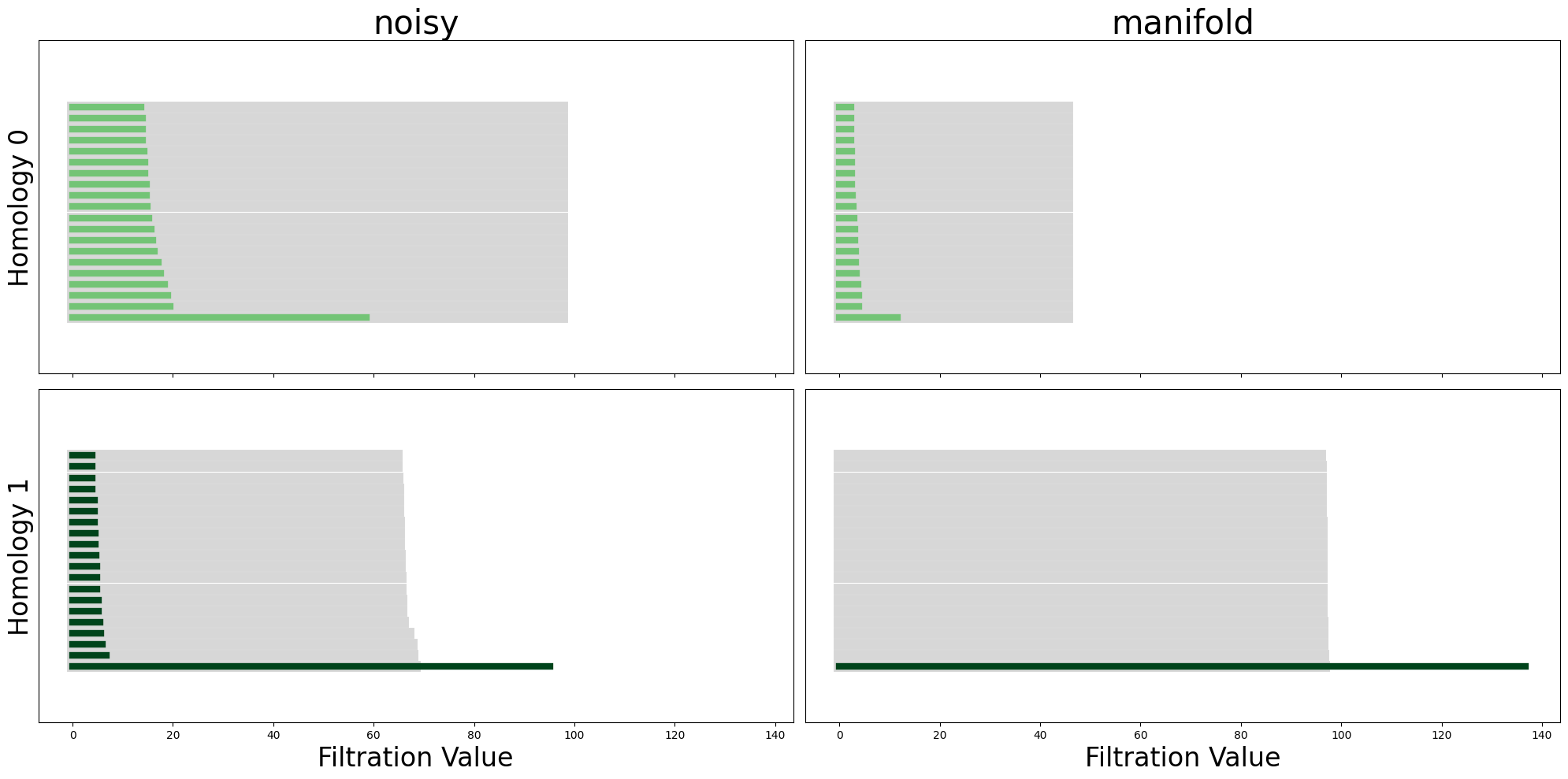
from neurometry.estimators.topology.plotting import plot_all_barcodes_with_null
plot_all_barcodes_with_null(
noisy_diagrams, "noisy", diagrams_2=manifold_diagrams, dataset_name_2="manifold"
);
In [10]:
def _get_lifespan_from_diagram(diagram):
birth = diagram[:, 0]
death = diagram[:, 1]
if np.isfinite(death).any():
inf_value = 3 * np.max(death[death != np.inf])
else:
inf_value = 1000
death[death == np.inf] = inf_value
lifespan = death - birth
indices = np.argsort(-lifespan)[:20]
return lifespan[indices]
In [11]:
def estimate_betti_numbers(points, num_shuffles, homology_dimensions=(0, 1, 2)):
all_diagrams = compute_diagrams_shuffle(
points, num_shuffles=num_shuffles, homology_dimensions=homology_dimensions
)
diagram = all_diagrams[0]
shuffled_diagrams = all_diagrams[1:]
betti_numbers = {dim: None for dim in homology_dimensions}
for dim in homology_dimensions:
filtered_diagram = diagram[diagram[:, 2] == dim]
lifespan = _get_lifespan_from_diagram(filtered_diagram)
filtered_shuffled_diagrams = np.array(
[
shuffled_diagrams[i, shuffled_diagrams[i, :, 2] == 1]
for i in range(shuffled_diagrams.shape[0])
]
)
betti_number = []
for diag in filtered_shuffled_diagrams:
shuffled_lifespan = _get_lifespan_from_diagram(diag)
significant_features = (lifespan > shuffled_lifespan).astype(int)
betti_number.append(sum(significant_features))
betti_numbers[dim] = (np.mean(betti_number), np.std(betti_number))
return betti_numbers
In [13]:
betti_numbers = estimate_betti_numbers(
manifold_points, num_shuffles=100, homology_dimensions=(0, 1, 2)
)
print(betti_numbers)
{0: (1.28, 2.209434316742636), 1: (1.0, 0.0), 2: (0.0, 0.0)}
In [67]:
homology_dimensions = (0, 1)
points = manifold_points
num_shuffles = 100
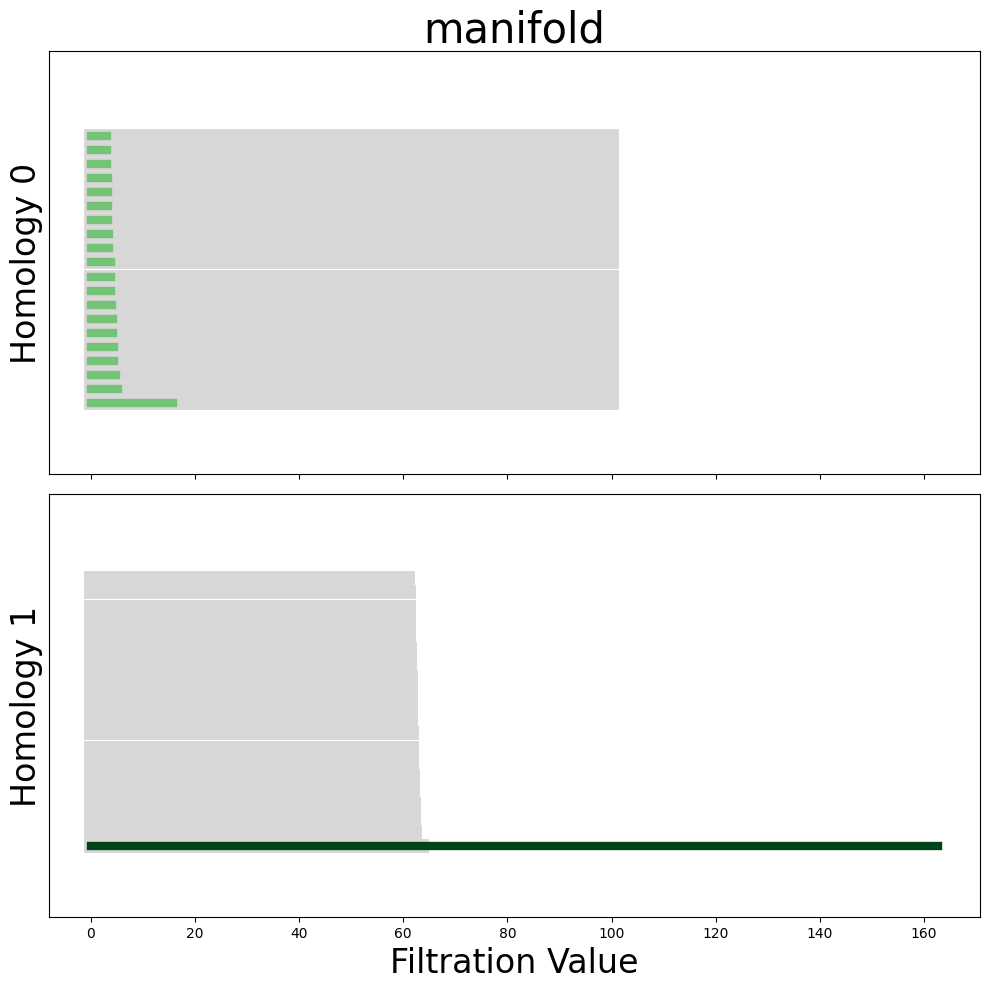
all_diagrams = compute_diagrams_shuffle(
points, num_shuffles=num_shuffles, homology_dimensions=homology_dimensions
)
plot_all_barcodes_with_null(all_diagrams, "manifold")
diagram = all_diagrams[0]
lifespan = _get_lifespan_from_diagram(diagram)
shuffled_diagrams = all_diagrams[1:]
betti_numbers = {dim: None for dim in homology_dimensions}
for dim in homology_dimensions:
betti_number = []
for shuffled_diagram in shuffled_diagrams:
shuffled_lifespan = _get_lifespan_from_diagram(shuffled_diagram)
significant_features = (lifespan > shuffled_lifespan).astype(int)
betti_number.append(sum(significant_features))
betti_numbers[dim] = np.mean(betti_number)
In [32]:
def get_betti_numbers(diagrams):
original_diagram = diagrams[0]
shuffled_diagrams = diagrams[1:]
dims = np.unique(original_diagram[:, 2]).astype(int)
betti_numbers = {dim: None for dim in dims}
for i, dim in enumerate(dims):
diagram_dim = original_diagram[original_diagram[:, 2] == dim]
null_diagram_dim = shuffled_diagrams[:, :, 2] == dim
null_diagram = shuffled_diagrams[null_diagram_dim]
null_lifespans_dim = _get_lifespan_from_diagram(null_diagram)
lifespans_dim = _get_lifespan_from_diagram(diagram_dim)
comparison = (lifespans_dim > null_lifespans_dim).astype(int)
betti_numbers[dim] = sum(comparison)
return betti_numbers
In [33]:
betti = get_betti_numbers(noisy_diagrams)
In [34]:
betti
Out [34]:
{0: 0, 1: 5, 2: 2}
Classify circle, sphere, torus point clouds#
In [68]:
import neurometry.datasets.synthetic as synthetic
from neurometry.estimators.topology.persistent_homology import compute_diagrams_shuffle
from gtda.diagrams import PersistenceEntropy
num_points = 1000
encoding_dim = 10
poisson_multiplier = 100
homology_dimensions = [0, 1, 2]
num_shuffles = 100
In [69]:
circle_task_points = synthetic.hypersphere(1, num_points)
circle_noisy_points, circle_manifold_points = synthetic.synthetic_neural_manifold(
points=circle_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=poisson_multiplier,
)
circle_noisy_diagrams = compute_diagrams_shuffle(
circle_noisy_points,
num_shuffles=num_shuffles,
homology_dimensions=homology_dimensions,
)
circle_PE = PersistenceEntropy()
circle_features = circle_PE.fit_transform(circle_noisy_diagrams)
noise level: 0.71%
In [70]:
sphere_task_points = synthetic.hypersphere(2, num_points)
sphere_noisy_points, sphere_manifold_points = synthetic.synthetic_neural_manifold(
points=sphere_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=poisson_multiplier,
)
sphere_noisy_diagrams = compute_diagrams_shuffle(
sphere_noisy_points,
num_shuffles=num_shuffles,
homology_dimensions=homology_dimensions,
)
sphere_PE = PersistenceEntropy()
sphere_features = sphere_PE.fit_transform(sphere_noisy_diagrams)
noise level: 0.71%
In [71]:
torus_task_points = synthetic.hypertorus(2, num_points)
torus_noisy_points, torus_manifold_points = synthetic.synthetic_neural_manifold(
points=torus_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=poisson_multiplier,
)
torus_noisy_diagrams = compute_diagrams_shuffle(
torus_noisy_points,
num_shuffles=num_shuffles,
homology_dimensions=homology_dimensions,
)
torus_PE = PersistenceEntropy()
torus_features = torus_PE.fit_transform(torus_noisy_diagrams)
noise level: 0.71%
In [72]:
# 3d plot with plotly
fig = go.Figure()
fig.add_trace(
go.Scatter3d(
x=circle_features[1:, 0],
y=circle_features[1:, 1],
z=circle_features[1:, 2],
mode="markers",
marker=dict(size=3, color="lightpink"),
name="Shuffled Circle",
)
)
fig.add_trace(
go.Scatter3d(
x=[circle_features[0, 0]],
y=[circle_features[0, 1]],
z=[circle_features[0, 2]],
mode="markers",
marker=dict(size=3, color="deeppink"),
name="Circle",
)
)
fig.add_trace(
go.Scatter3d(
x=sphere_features[1:, 0],
y=sphere_features[1:, 1],
z=sphere_features[1:, 2],
mode="markers",
marker=dict(size=3, color="lightgreen"),
name="Shuffled Sphere",
)
)
fig.add_trace(
go.Scatter3d(
x=[sphere_features[0, 0]],
y=[sphere_features[0, 1]],
z=[sphere_features[0, 2]],
mode="markers",
marker=dict(size=3, color="darkgreen"),
name="Sphere",
)
)
fig.add_trace(
go.Scatter3d(
x=torus_features[1:, 0],
y=torus_features[1:, 1],
z=torus_features[1:, 2],
mode="markers",
marker=dict(size=3, color="lightblue"),
name="Shuffled Torus",
)
)
fig.add_trace(
go.Scatter3d(
x=[torus_features[0, 0]],
y=[torus_features[0, 1]],
z=[torus_features[0, 2]],
mode="markers",
marker=dict(size=3, color="darkblue"),
name="Torus",
)
)
In [16]:
num_points = 500
encoding_dim = 10
poisson_multiplier = 100
test_task_points = synthetic.hypertorus(2, 400)
test_noisy_points, test_manifold_points = synthetic.synthetic_neural_manifold(
points=test_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=poisson_multiplier,
)
from neurometry.estimators.topology.persistent_homology import TopologicalClassifier
num_samples = 10
TC = TopologicalClassifier(
num_samples=num_samples, poisson_multiplier=poisson_multiplier, reduce_dim=True
)
TC.fit(test_noisy_points)
Classifier score: 1.0
Out [16]:
TopologicalClassifier(num_samples=10, poisson_multiplier=100, reduce_dim=True)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
TopologicalClassifier(num_samples=10, poisson_multiplier=100, reduce_dim=True)
In [17]:
TC.predict(test_noisy_points)
Out [17]:
array([2.])
In [8]:
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.base import BaseEstimator, ClassifierMixin
class TopologicalClassifier(ClassifierMixin, BaseEstimator):
def __init__(
self,
num_samples,
poisson_multiplier,
homology_dimensions=[0, 1, 2],
reduce_dim=False,
):
self.num_samples = num_samples
self.poisson_multiplier = poisson_multiplier
self.homology_dimensions = homology_dimensions
self.reduce_dim = reduce_dim
self.classifier = RandomForestClassifier()
def _generate_ref_data(self, input_data):
num_points = input_data.shape[0]
encoding_dim = input_data.shape[1]
circle_task_points = synthetic.hypersphere(1, num_points)
circle_point_clouds = []
for i in range(self.num_samples):
circle_noisy_points, _ = synthetic.synthetic_neural_manifold(
points=circle_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=self.poisson_multiplier,
)
circle_point_clouds.append(circle_noisy_points)
sphere_task_points = synthetic.hypersphere(2, num_points)
sphere_point_clouds = []
for i in range(num_samples):
sphere_noisy_points, _ = synthetic.synthetic_neural_manifold(
points=sphere_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=self.poisson_multiplier,
)
sphere_point_clouds.append(sphere_noisy_points)
torus_task_points = synthetic.hypertorus(2, num_points)
torus_point_clouds = []
for i in range(num_samples):
torus_noisy_points, _ = synthetic.synthetic_neural_manifold(
points=torus_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=self.poisson_multiplier,
)
torus_point_clouds.append(torus_noisy_points)
klein_bottle_task_points = synthetic.klein_bottle(num_points)
klein_bottle_point_clouds = []
for i in range(num_samples):
klein_bottle_noisy_points, _ = synthetic.synthetic_neural_manifold(
points=klein_bottle_task_points,
encoding_dim=encoding_dim,
nonlinearity="sigmoid",
scales=gs.ones(encoding_dim),
poisson_multiplier=self.poisson_multiplier,
)
klein_bottle_point_clouds.append(klein_bottle_noisy_points)
circle_labels = np.zeros(num_samples)
sphere_labels = np.ones(num_samples)
torus_labels = 2 * np.ones(num_samples)
klein_bottle_labels = 3 * np.ones(num_samples)
ref_labels = np.concatenate(
[
circle_labels,
sphere_labels,
torus_labels,
klein_bottle_labels,
]
)
ref_point_clouds = [
*circle_point_clouds,
*sphere_point_clouds,
*torus_point_clouds,
*klein_bottle_point_clouds,
]
return ref_point_clouds, ref_labels
def _compute_topo_features(self, diagrams):
PE = PersistenceEntropy()
features = PE.fit_transform(diagrams)
return features
def fit(self, X, y=None):
ref_point_clouds, ref_labels = self._generate_ref_data(X)
if self.reduce_dim:
pca = PCA(n_components=10)
ref_point_clouds = [
pca.fit_transform(point_cloud) for point_cloud in ref_point_clouds
]
ref_diagrams = compute_persistence_diagrams(
ref_point_clouds, homology_dimensions=self.homology_dimensions
)
ref_features = self._compute_topo_features(ref_diagrams)
X_ref_train, X_ref_valid, y_ref_train, y_ref_valid = train_test_split(
ref_features, ref_labels
)
self.classifier.fit(X_ref_train, y_ref_train)
print(f"Classifier score: {self.classifier.score(X_ref_valid, y_ref_valid)}")
return self
def predict(self, X):
if self.reduce_dim:
pca = PCA(n_components=10)
X = pca.fit_transform(X)
diagram = compute_persistence_diagrams(
[X], homology_dimensions=self.homology_dimensions
)
features = self._compute_topo_features(diagram)
return self.classifier.predict(features)
In [8]:
test_task_points = synthetic.hypertorus(2, 400)
test_noisy_points, test_manifold_points = synthetic.synthetic_neural_manifold(
points=test_task_points,
encoding_dim=5,
nonlinearity="sigmoid",
scales=gs.ones(10),
poisson_multiplier=100,
)
TC = TopologicalClassifier(num_samples=10, poisson_multiplier=100, reduce_dim=True)
TC.fit(test_noisy_points)
In [116]:
# Example color mapping
color_map = {0: "darkblue", 1: "deeppink", 2: "limegreen", 3: "orange"}
colors = [color_map[label] for label in labels]
names = {0: "Circle", 1: "Sphere", 2: "Torus", 3: "Klein Bottle"}
# Create a figure
fig = go.Figure()
# Add the trace with colored markers based on labels
fig.add_trace(
go.Scatter3d(
x=features[:, 0],
y=features[:, 1],
z=features[:, 2],
mode="markers",
marker=dict(size=3, color=colors),
showlegend=False,
)
)
# Manually add the legend entries
for label, color in color_map.items():
fig.add_trace(
go.Scatter3d(
x=[None],
y=[None],
z=[None],
mode="markers",
marker=dict(size=3, color=color),
showlegend=True,
name=f"{names[label]}",
)
)
fig.show()
In [110]:
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(features, labels)
model = RandomForestClassifier()
model.fit(X_train, y_train)
model.score(X_valid, y_valid)
Out [110]:
0.8
In [9]:
test_task_points = synthetic.hypertorus(2, 400)
test_noisy_points, test_manifold_points = synthetic.synthetic_neural_manifold(
points=test_task_points,
encoding_dim=5,
nonlinearity="sigmoid",
scales=gs.ones(10),
poisson_multiplier=100,
)
TC.fit(test_noisy_points)
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
Cell In[9], line 2
1 test_task_points = synthetic.hypertorus(2, 400)
----> 2 test_noisy_points, test_manifold_points = synthetic.synthetic_neural_manifold(
3 points=test_task_points,
4 encoding_dim=5,
5 nonlinearity="sigmoid",
6 scales=gs.ones(10),
7 poisson_multiplier=100,
8 )
10 TC.fit(test_noisy_points)
File ~/neurometry/neurometry/datasets/synthetic.py:42, in synthetic_neural_manifold(points, encoding_dim, nonlinearity, poisson_multiplier, ref_frequency, **kwargs)
40 encoding_matrix = random_encoding_matrix(manifold_extrinsic_dim, encoding_dim)
41 encoded_points = encode_points(points, encoding_matrix)
---> 42 manifold_points = ref_frequency * apply_nonlinearity(
43 encoded_points, nonlinearity, **kwargs
44 )
45 try:
46 noisy_points = poisson_spikes(manifold_points, poisson_multiplier)
File ~/neurometry/neurometry/datasets/synthetic.py:218, in apply_nonlinearity(encoded_points, nonlinearity, **kwargs)
216 return relu(encoded_points, **kwargs)
217 if nonlinearity == "sigmoid":
--> 218 return scaled_sigmoid(encoded_points, **kwargs)
219 if nonlinearity == "tanh":
220 return scaled_tanh(encoded_points, **kwargs)
File ~/neurometry/neurometry/datasets/synthetic.py:231, in scaled_sigmoid(tensor, scales)
230 def scaled_sigmoid(tensor, scales):
--> 231 assert tensor.shape[1] == scales.shape[0], "scales must have same shape as tensor"
232 return 1 / (1 + gs.exp(-scales * tensor))
AssertionError: scales must have same shape as tensor
NOTE: Try different coefficients to distinguish between torus and klein bottle?#
In [105]:
betti_numbers = estimate_betti_numbers(
manifold_points, num_shuffles=500, homology_dimensions=(0, 1)
)
print(betti_numbers)
{0: (19.814, 0.7066852198822331), 1: (13.214, 2.555817677378416)}
In [12]:
num_shuffles = 10
noisy_diagrams = compute_diagrams_shuffle(
noisy_points, num_shuffles=num_shuffles, homology_dimensions=(0, 1, 2)
)
manifold_diagrams = compute_diagrams_shuffle(
manifold_points, num_shuffles=num_shuffles, homology_dimensions=(0, 1, 2)
)
plot_all_barcodes_with_null(
noisy_diagrams, "noisy", diagrams_2=manifold_diagrams, dataset_name_2="manifold"
);
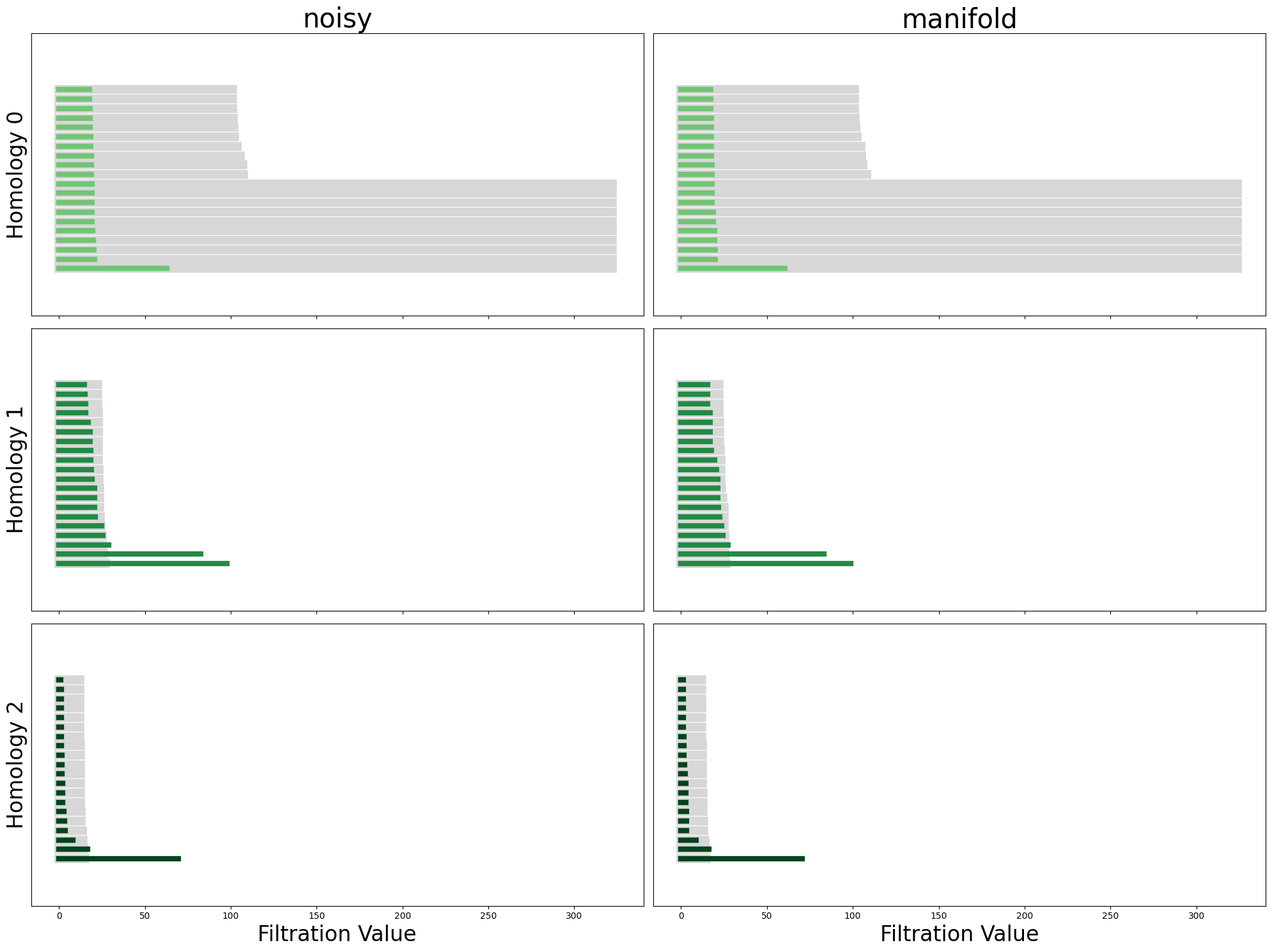
In [13]:
betti = get_betti_numbers(noisy_diagrams)
print(betti)
{0: 0, 1: 5, 2: 2}
TODO: FIX get_betti_numbers FUNCTION!!!!#
Also: how to deal with betti 0 ? There should be a single “significant” feature?
In [14]:
sphere_1_betti = {0: 1, 1: 1, 2: 0}
torus_2_betti = {0: 1, 1: 2, 2: 1}
sphere_2_betti = {0: 1, 1: 0, 2: 1}
# if betti == sphere_1_betti:
# print("The manifold is a ring") ----> use ring VAE
# elif betti == torus_2_betti: