Notebook source code: notebooks/04_explore_hyperbolic_geometry.ipynb
Explore Hyperbolic Geometry#
Imports & Setup#
In [13]:
import setup
setup.main()
import os
os.environ["GEOMSTATS_BACKEND"] = "pytorch"
import geomstats.backend as gs
import matplotlib.pyplot as plt
import numpy as np
from geomstats.geometry.hyperboloid import Hyperboloid
from geomstats.geometry.pullback_metric import PullbackDiffeoMetric, PullbackMetric
from viz import plot_grids
import neurometry.datasets.gridcells as gridcells
%load_ext autoreload
%autoreload 2
%matplotlib inline
Working directory: /Users/facosta/Desktop/code/neurometry/neurometry/neuralwarp
Directory added to path: /Users/facosta/Desktop/code/neurometry/neurometry
Directory added to path: /Users/facosta/Desktop/code/neurometry/neurometry/neuralwarp
['/Users/facosta/Desktop/code/neurometry/neurometry/neuralwarp', '/Users/facosta/miniconda3/envs/neurometry/lib/python38.zip', '/Users/facosta/miniconda3/envs/neurometry/lib/python3.8', '/Users/facosta/miniconda3/envs/neurometry/lib/python3.8/lib-dynload', '', '/Users/facosta/miniconda3/envs/neurometry/lib/python3.8/site-packages', '/Users/facosta/Desktop/code/neurometry', '/Users/facosta/Desktop/code/neurometry/neurometry', '/Users/facosta/Desktop/code/neurometry/neurometry/neuralwarp', '/Users/facosta/Desktop/code/neurometry', '/Users/facosta/Desktop/code/neurometry/neurometry', '/Users/facosta/Desktop/code/neurometry/neurometry/neuralwarp']
Hyperbolic Geometry#
In [14]:
H2 = Hyperboloid(dim=2)
def convert_to_poincare_coordinates(points):
return points[1:] / (1 + points[:1])
def hyperbolic_warp(point):
extrinsic_point = H2.from_coordinates(point, "intrinsic")
return convert_to_poincare_coordinates(extrinsic_point)
warp = hyperbolic_warp
In [16]:
# old geomstats
pullback_metric = PullbackMetric(dim=2, embedding_dim=2, immersion=warp)
In [15]:
# new geomstats
class PlaneWarpMetric(PullbackDiffeoMetric):
def __init__(self):
super().__init__(space=Euclidean(dim=2))
def _define_embedding_space(self):
return Euclidean(dim=2)
def diffeomorphism(self, base_point):
return warp(base_point)
def inverse_diffeomorphism(self, base_point):
return warp(base_point)
pullback_metric = PlaneWarpMetric()
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[15], line 12
9 def inverse_diffeomorphism(self, base_point):
10 return warp(base_point)
---> 12 pullback_metric = PlaneWarpMetric()
TypeError: Can't instantiate abstract class PlaneWarpMetric with abstract methods define_embedding_metric
In [5]:
point_a = gs.array([0.0, 0.0])
point_b = gs.array([1.0, 1.0])
pullback_metric.dist(point_a, point_b)
Out [5]:
tensor(0.5221)
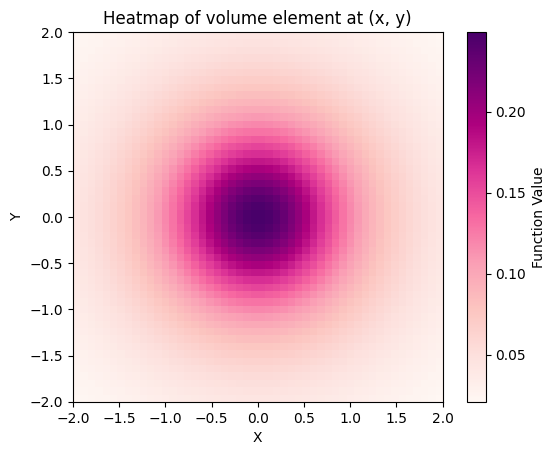
Plot volume element \(\sqrt{|\text{det}(g)|}\)#
In [7]:
x = gs.linspace(-2, 2, 50)
y = gs.linspace(-2, 2, 50)
x_grid, y_grid = gs.meshgrid(x, y)
# Combine and reshape the x and y coordinates into a list of 2D points
points = gs.vstack((x_grid.ravel(), y_grid.ravel())).T
# Define your function
def volume_element(x, y):
point = gs.array([x, y])
g = pullback_metric.metric_matrix(point)
return gs.sqrt(gs.abs(gs.linalg.det(g)))
# Apply the function to each point in the list
values = gs.array([volume_element(x, y) for x, y in points])
# Reshape the values back into a 2D grid
z_values = values.reshape(x_grid.shape)
# Create the heatmap
plt.imshow(
z_values, origin="lower", extent=[x.min(), x.max(), y.min(), y.max()], cmap="RdPu"
)
plt.colorbar(label="Function Value")
plt.xlabel("X")
plt.ylabel("Y")
plt.title("Heatmap of volume element at (x, y)")
plt.show()
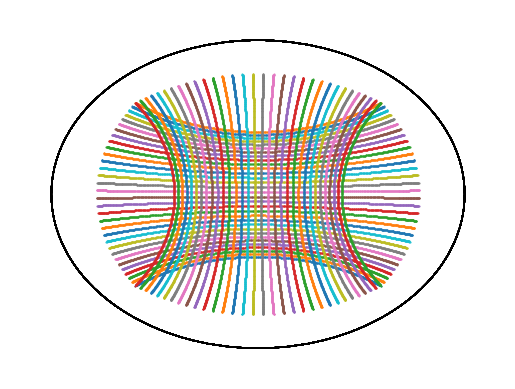
Visualize geodesic grid#
In [11]:
"""Plot a grid on H2 with Poincare Disk visualization."""
import geomstats.backend as gs
import geomstats.visualization as visualization
import matplotlib.pyplot as plt
from geomstats.geometry.hyperboloid import Hyperboloid
H2 = Hyperboloid(dim=2)
METRIC = H2.metric
# from geomstats.geometry.euclidean import Euclidean
# R2 = Euclidean(dim=2)
# METRIC = R2.metric
left = -4.0
right = 4.0
bottom = -4.0
top = 4.0
grid_size = 32
n_steps = 512
"""Plot a grid on H2 with Poincare Disk visualization.
Parameters
----------
left, right, bottom, top : ints
Grid's coordinates
grid_size : int
Grid's size.
n_steps : int
Number of steps along the geodesics defining the grid.
"""
starts = []
ends = []
for p in gs.linspace(left, right, grid_size):
starts.append(gs.array([top, p]))
ends.append(gs.array([bottom, p]))
for p in gs.linspace(top, bottom, grid_size):
starts.append(gs.array([p, left]))
ends.append(gs.array([p, right]))
starts = [H2.from_coordinates(s, "intrinsic") for s in starts]
ends = [H2.from_coordinates(e, "intrinsic") for e in ends]
ax = plt.gca()
for start, end in zip(starts, ends, strict=False):
geodesic = METRIC.geodesic(initial_point=start, end_point=end)
t = gs.linspace(0.0, 1.0, n_steps)
points_to_plot = geodesic(t)
visualization.plot(points_to_plot, ax=ax, space="H2_poincare_disk", marker=".", s=1)
In [ ]:
from geomstats.geometry.hyperboloid import Hyperboloid
H2 = Hyperboloid(dim=2)
def hyperbolic_wrap(s):
return H2.from_coordinates(s, "intrinsic")
In [ ]:
visualization.plot(points_to_plot, ax=ax, space="H2_poincare_disk", marker=".", s=1)
In [ ]:
from geomstats.geometry.poincare_ball import PoincareBall
H2 = PoincareBall(dim=2)
def hyperbolic_wrap(point):
return H2.change_coordinates_system(
point, from_coordinates_system="intrinsic", to_coordinates_system="ball"
)
In [ ]:
from geomstats.geometry.hyperboloid import Hyperboloid
H2 = Hyperboloid(dim=2) # extrinsic coordinates
def convert_to_poincare_coordinates(points):
return points[1:] / (1 + points[:1])
# def convert_to_klein_coordinates(points):
# poincare_coords = points[:, 1:] / (1 + points[:, :1])
# poincare_radius = gs.linalg.norm(poincare_coords, axis=1)
# poincare_angle = gs.arctan2(poincare_coords[:, 1], poincare_coords[:, 0])
# klein_radius = 2 * poincare_radius / (1 + poincare_radius**2)
# klein_angle = poincare_angle
# coords_0 = gs.expand_dims(klein_radius * gs.cos(klein_angle), axis=1)
# coords_1 = gs.expand_dims(klein_radius * gs.sin(klein_angle), axis=1)
# klein_coords = gs.concatenate([coords_0, coords_1], axis=1)
# return klein_coords
def convert_to_klein_coordinates(points):
"""Convert from extrinsic to klein coordinates."""
poincare_coords = points[1:] / (1 + points[:1])
poincare_radius = gs.linalg.norm(poincare_coords)
poincare_angle = gs.arctan2(poincare_coords[1], poincare_coords[0])
klein_radius = 2 * poincare_radius / (1 + poincare_radius**2)
klein_angle = poincare_angle
coords_0 = klein_radius * gs.cos(klein_angle)
coords_1 = klein_radius * gs.sin(klein_angle)
return gs.array([coords_0, coords_1])
def hyperbolic_wrap(point):
"""Hyperbolic wrap happens in two steps:
- convert from intrinsic to extrinsic
- convert from extrinsic to klein/poincare**
"""
extrinsic_point = H2.from_coordinates(point, "intrinsic")
return convert_to_klein_coordinates(extrinsic_point)
Plot grids on hyperbolic space#
In [17]:
grids, grids_warped = gridcells.generate_all_grids(
grid_scale,
lx,
ly,
arena_dims,
n_cells,
grid_orientation_mean,
grid_orientation_std,
warp=hyperbolic_wrap,
lattice_type="square",
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[17], line 2
1 grids, grids_warped = gridcells.generate_all_grids(
----> 2 grid_scale, lx, ly, arena_dims, n_cells, grid_orientation_mean, grid_orientation_std, warp=hyperbolic_wrap, lattice_type = "square")
NameError: name 'lx' is not defined
In [ ]:
plot_grids(grids, arena_dims)
plot_grids(grids_warped, arena_dims)
In [9]:
# import neurometry.datasets.structures as structures
import neurometry.datasets.gridcells as gridcells
scale = 1
lattice_type = "hexagonal"
dimensions = np.array([4, 4])
n_cells = 1
# lattice = structures.get_lattice(scale,lattice_type,dimensions)
In [12]:
grids, grids_warped = gridcells.generate_all_grids(
scale, dimensions, n_cells, warp=hyperbolic_warp, lattice_type="square"
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[12], line 1
----> 1 grids, grids_warped = gridcells.generate_all_grids(scale, dimensions, n_cells, warp=hyperbolic_warp, lattice_type = "square")
File ~/Desktop/code/neurometry/neurometry/datasets/gridcells.py:111, in generate_all_grids(grid_scale, arena_dims, n_cells, grid_orientation_mean, grid_orientation_std, warp, lattice_type)
105 angle_i = np.random.normal(grid_orientation_mean, grid_orientation_std) * (
106 np.pi / 180
107 )
108 rot_i = np.array(
109 [[np.cos(angle_i), -np.sin(angle_i)], [np.sin(angle_i), np.cos(angle_i)]]
110 )
--> 111 phase_i = np.multiply([lx, ly], np.random.rand(2))
112 lattice_i = np.matmul(rot_i, ref_lattice.T).T + phase_i
113 #lattice_i = np.where(abs(lattice_i) < arena_dims / 2, lattice_i, None)
NameError: name 'lx' is not defined
In [ ]: